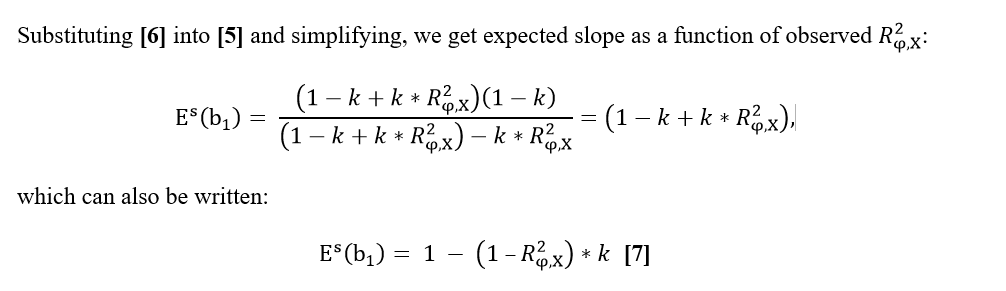|
Size: 48444
Comment: Instructions from Valentina
|
Size: 22894
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from public/CoPAppendixX {{attachment:CoP_banner2.png}} |
#pragma section-numbers = Interbull CoP - Appendix VIII - Interbull validation test for genomic evaluations - GEBV test = ''Document based on: '' |
| Line 4: | Line 5: |
| = Appendix VIII - Interbull validation test for genomic evaluations – GEBV test = ''Document based on Mäntysaari, E., Liu, Z and VanRaden P. 2011. Interbull Validation Test for Genomic Evaluations. Interbull Bulletin 41, p. 17-21.'' |
''Sullivan, P.G. 2023. Updated Interbull software for genomic validation tests. Interbull Bulletin 58, p.7-16. '' |
| Line 7: | Line 7: |
| . <<TableOfContents>> | ''!VanRaden, P.M. 2021. Improved genomic validation including extra regressions. Interbull bulletin 56: 65-69.'' ''Mäntysaari, E., Liu, Z and !VanRaden P. 2010. Interbull Validation Test for Genomic Evaluations. Interbull Bulletin 41, p. 17-21.'' |
| Line 10: | Line 12: |
| * EBV – Estimated Breeding Value (conventional national evaluations of the trait, free of genomic information, which are submitted to Interbull to be used in MACE evaluations) * DGV - Direct Estimated Genomic Value (genomic evaluations based on SNP prediction equations) * GEBV – Genomically Enhanced Estimated Breeding Value (evaluations that combine EBV and DGV) * EDC –Effective Daughter Contribution * GEDC – Genomically Enhanced Effective Daughter Contribution (EDC plus the genomic contribution) * GMACE - Multiple Trait Across Country Genomic Evaluation * PA – Parent Average * D_PGM – De-regressed Predicted Genetic Merit * DD – Daughter Deviation * NGEC - National Genetic Evaluation Centre * λ = (4-h^2^)/h^2^ * r^2^ – Reliability of the bull’s evaluation * R^2^ – Accuracy of the test model ''' ''' |
1. EBV - Estimated Breeding Value (conventional national evaluations of the trait, free of genomic information, which are submitted to Interbull to be used in MACE evaluations) 1. DGV - Direct Estimated Genomic Value (genomic evaluations based on SNP prediction equations) 1. GEBV - Genomically Enhanced Estimated Breeding Value (evaluations that combine EBV and DGV) 1. dGEBV - De-regressed GEBV 1. GREL - Genomic reliability of the bull's GEBV 1. EDC - Effective Daughter Contribution 1. MACE - Multiple Trait Across Country Evaluation 1. PA - Parent Average 1. DD - Daughter Deviation 1. NGEC - National Genetic Evaluation Centre |
| Line 25: | Line 33: |
| The inclusion of genomic information in international comparisons for dairy breeds requires that the national genomic breeding values (GEBVs) get validated by Interbull in a similar fashion that conventional EBVs are validated as a pre-condition to participate in the MACE evaluations. | The inclusion of national genomic information in international comparisons for dairy breeds requires that the national genomic breeding values (GEBV) get validated by Interbull in a similar fashion that conventional EBV are validated as a pre-condition to participate in the MACE evaluations. |
| Line 27: | Line 35: |
| The '''''GEBV test''''' will be applied to validate national models used to compute GEBVs that the national genetic evaluation centers (NGEC) publish and will eventually submit to Interbull for international genetic evaluations including genomic information. The '''''GEBV test''''' can be considered also a quality assurance assessment for national genomic evaluations. GEBVs from models that have been tested can be referred to as breeding value estimates with appropriate reliability, and be converted to other country scale breeding values using conversion equations derived by Interbull. | The '''''GEBV test''''' will be applied to validate national models used to compute GEBV that the national genetic evaluation centers (NGEC) publish and will eventually submit to Interbull for international genetic evaluations including genomic information. The ''GEBV test'' can also be considered a quality assurance assessment for national genomic evaluations. GEBV from models that have been tested can be referred to as breeding value estimates with appropriate reliability, and which can be converted to other country scale breeding values using conversion equations derived by Interbull. |
| Line 30: | Line 38: |
| The '''''GEBV test''''' evaluates: | The '''GEBV test''' evaluates: |
| Line 32: | Line 40: |
| * the unbiasedness of the genomic evaluations through the evaluation of * the consistency of the genetic trend captured by GEBV, and * the consistency of the variation of GEBVs and EBVs; * the improvement in accuracy from the use of GEBV instead of EBV. |
1. the unbiasedness of the genomic evaluations through the evaluation of 1. the consistency of the genetic trend captured by GEBV, 1. the consistency of bull rankings before versus after having progeny, and 1. the consistency of the variation of GEBV relative to EBV; |
| Line 37: | Line 45: |
| The test for bias is done by verifying the ability of a model only including data from 4 years ago to predict current performances. NGEC have to exclude the last 4 years of data and re-run the analyses with the reduced data, with the same model that are being tested. However, in some cases the bull generation available for validation has not been genotyped in everything and all. Thus, bulls exist that will get more than 20 daughters in the full data, but that have no GEBVs. This is called selective genotyping, and it leads into systematic bias in the validation bull group. In the test, this bias needs to be corrected by accounting for the selection between the mean national EBV (current, conventional) of the bulls genotyped and the overall mean national EBV including all potential candidates. This selection differential can be used to derive the expected regression coefficient, which would be equal to unity as if no selective genotyping took place. <<BR>>Testing the improvement in accuracy is done by comparing the coefficient of determination (R^2^) of the reduced genomic model and the equivalent reduced conventional model (from 4 years ago) regressed to current performances. The R^2^ from the model including genomic information must be higher than the model including only parent average information. | 1. the improvement in selection accuracy from the use of GEBV instead of EBV. A time-oriented cross-validation is used to test how well genomic evaluations of young bull calves, using current models and phenotypic data from 4 years ago, can predict current progeny performance. The NGEC shall re-run their current evaluation software while excluding the most recent 4 years of daughter phenotypes, to obtain reduced-data genetic (EBVr) and genomic (GEBVr) evaluations. The software will then test if the ranking and variance of bull GEBVr match statistical and genetic expectations relative to ranking and variance of the bull comparisons based on current progeny differences, as an indication of unbiasedness. Furthermore, if the GEBVr are more highly correlated than EBVr with the current progeny phenotypes, it is an indication of accuracy improvement with GEBV. Linear regression models are used for the validation test, where the expected value of regression slopes equals 1 if validation bulls are an unselected group, and a value less than 1 if only a selected subgroup of the most recent proven bulls have been genotyped. The expected slope is lower with selective genotyping due to effects of selection on variances and covariances used to compute the validation slope. The software will account for effects of selective genotyping on expected slopes, using estimates of selection differential from the differences between average EBV of the genotyped bulls versus all proven bulls born in the period considered for validation testing. Bootstrapping is used for all significance testing, and a combination of statistical and biological limits of tolerance is used by Interbull to assign an overall assessment of pass or fail. |
| Line 40: | Line 52: |
| Data formats are described at '''[[https://wiki.interbull.org/public/GEBVtest_software?action=print|GEBVtest Software]]'''. | Data formats are described at https://interbull.org/ib/gebvtest_software_2024 |
| Line 42: | Line 54: |
| === Full data sets === The full data sets include all animals present in the most recent Interbull MACE evaluation. They are of two types, one containing national official genetic merit values (EBVs) and another containing either de-regressed predicted genetic merits (D_PGMs) or daughter deviations (DDs). |
== Full data sets == Two sets of currently official evaluations for progeny-proven bulls shall be provided for the GEBV test. These will be the EBV and GEBV published or otherwise indirectly used by the NGEC for national selection programs. All bulls provided to Interbull in file300 for MACE shall be included in a conventional EBV file (file300Cf) for the GEBV test, and all these same bulls who are genotyped and have a national GEBV shall be included in the GEBV file (file300Gf). |
| Line 45: | Line 57: |
| ==== National official genetic evaluation file (fileCxxxf) ==== The files sent by the NGEC as input for the most recent Interbull MACE evaluation and will be used to identify the candidate bulls, estimate selection intensity and check bulls birth year and type of proof. |
* '''Conventional national genetic evaluation file (file300Cf)''' |
| Line 48: | Line 59: |
| ==== Daughter deviation file (fileDxxxf) ==== The NGEC needs to prepare either DD or D_PGM for the same animals included in fileCxxxf. These values represent the currently estimated performance of the animals and will be used as the dependent variable in the validation procedure. EDC and reliability estimates should be exactly the same as in fileCxxxf. |
The national EBV sent by the NGEC as input for the most recent Interbull MACE evaluation will be used to identify validation test candidate bulls, estimate the intensity of selective genotyping, and check bulls birth year and type of proof. |
| Line 51: | Line 61: |
| === Reduced data sets === The reduced data sets should be prepared by truncating the phenotypes used as input for both the conventional and the genomic evaluations. The NGEC must exclude phenotypic information from the past 4 years and re-run the current models of genetic/genomic evaluation for the traits of interest, keeping the animals without progeny information after truncation (test bulls) in the data in order to obtain genetic merit estimates based solely on parent averages (EBVr) or on parent averages plus genomic prediction equations (GEBVr). |
* '''Official national genomic evaluation file (file300Gf)''' |
| Line 54: | Line 63: |
| ==== Reduced conventional genetic evaluation file (fileCxxxr) ==== The NGEC should carry out a conventional genetic evaluation using truncated data (only phenotypes up to 4 years prior to the date of analysis) but including in the analysis all animals present in the current official evaluations (fileCxxxf).--( )-- |
The national GEBV of current MACE bulls will be used to derive target values reflecting unbiased estimates of average progeny performance for the validation test bulls. The official validation target is derived internally by the software, based on the consistent application for all NGEC of a standardized international method for dGEBV developed by !VanRaden(2021). |
| Line 57: | Line 65: |
| ==== Reduced genomic evaluation file (fileGxxxr) ==== Similarly, new genomic evaluations should be carried out using exactly the same model being validated (current) but excluding phenotypic information up to four years ago (truncated data, fileCxxxr). All bulls that did not have a progeny test 4 years ago and that currently have at least 20 daughter-equivalents in the national genetic evaluation (test bulls) need to have a genomically enhanced EBV (GEBVr) estimated and included in the output. |
* '''Reduced data sets''' |
| Line 60: | Line 67: |
| If a significant number of foreign animals are included in the reference population and estimation of genomic prediction equations uses de-regressed MACE values for these animals as input, the reduced genomic evaluation can be achieved in two ways: | The reduced data sets should be prepared by truncating the phenotypes used as input for both the conventional and the genomic evaluations. The NGEC must exclude phenotypic information from the most recent 4 years and re-run the current models of genetic and genomic evaluation for the traits of interest. The pedigree should not be truncated, just the phenotypes, because each validation bull's predicted genetic contributions in future progeny, based solely on the bull's parent average (EBVr=PA) and on PA plus genomic prediction equations (GEBVr) from the reduced-data evaluations will be needed for the validation test. |
| Line 62: | Line 69: |
| a. the Interbull Centre can make historical files available upon request (e.g. information used four years ago) containing past MACE results and the correspondent national EDCs, as well as heritability and genetic correlations used in the respective evaluations – these data can then be used to estimate 4-year old de-regressed values; OR a. the genomic prediction equations for the truncated data (only bulls with EDCr > 0) are obtained using current de-regressed MACE values. This constitutes an exception and should only be used when the standard procedure is not practical. |
* '''Reduced conventional genetic evaluation file (file300Cr)''' |
| Line 65: | Line 71: |
| Table 1 presents a comparison between the several types of data and the notation used to identify variables from different files. | The NGEC shall carry out a conventional genetic evaluation with no genotypes, while using the truncated data (only phenotypes up to 4 years prior to the date of analysis) but including in the analysis all animals present in the current official evaluations used in MACE (file300Cf). |
| Line 67: | Line 73: |
| '''Table 1 –''' Comparative specification of the data files needed for the '''''GEBV test'''''. ''' ''' ||<tablestyle="margin-left:-5.3pt;border-collapse:collapse;border:none;mso-border-alt:solid windowtext .5pt;mso-yfti-tbllook:160;mso-padding-alt:0cm 5.4pt 0cm 5.4pt;mso-border-insideh:.5pt solid windowtext;mso-border-insidev:.5pt solid windowtext" tableclass="MsoNormalTable"rowstyle="mso-yfti-irow:0;mso-yfti-firstrow:yes"#D9D9D9 height="6.75pt" style="border:solid windowtext 1.0pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;text-align:center" |2>Test Data ''' ''' ||<#D9D9D9 height="6.75pt" style="border:solid windowtext 1.0pt;border-left:none;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;text-align:center" |2>Type of information ''' ''' ||<#D9D9D9 height="6.75pt" style="border:solid windowtext 1.0pt;border-left:none;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;text-align:center" |2>File types and format^a^ ''' ''' ||||||<#D9D9D9 height="6.75pt" style="border:solid windowtext 1.0pt;border-left:none;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;text-align:center;vertical-align:top">Specific variables^b^ '''(equivalent field in the fileCxxxf)''' ''' ''' || ||<rowstyle="mso-yfti-irow:1"#D9D9D9 height="6.75pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;vertical-align:top">EDC ||<#D9D9D9 height="6.75pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;vertical-align:top">Reliability ||<#D9D9D9 height="6.75pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;vertical-align:top">EBV || ||<rowstyle="mso-yfti-irow:2"style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;text-align:center" |2>Full data sets ''' ''' ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">Conventional Genetic data ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">C010f, C115f, C015f, C016f, C017f, C018f, C019f, C020f ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">EDC ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">r^2^,,EBV,, ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">EBV || ||<rowstyle="mso-yfti-irow:3"style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">Daughter deviation data ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">D010f, D115f, D015f, D016f, D017f, D018f, D019f, D020f ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">EDC ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">r^2^,,EBV,, ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">D_PGM (or DD, if available) || ||<rowstyle="mso-yfti-irow:4"style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt;text-align:center" |2>Reduced data sets ''' ''' ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">Conventional Genetic data ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">C010r, C115r, C015r, C016r, C017r, C018r, C019r, C020r ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">EDCr ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">r^2^,,EBVr,, ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">EBVr || ||<rowstyle="mso-yfti-irow:5;mso-yfti-lastrow:yes"style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">Genomic data ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">G010r, G115r, G015r, G016r, G017r, G018r, G019r, G020r ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">GEDCr ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">r^2^,,GEBVr,, ||<style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt">GEBVr || |
A minimum of 10 most recent birth years of proven bulls included in file300Cf must also be included in file300Cr. The older proven bulls, with progeny proofs already in the reduced data, are required as a comparative control group, to contrast evaluation changes for younger bulls in the validation test group relative to the older control bulls. |
| Line 75: | Line 75: |
| * '''Reduced genomic evaluation file (file300Gr)''' | |
| Line 76: | Line 77: |
| The NGEC shall carry out a genomic evaluation that includes the genotypes, while using the truncated data (only phenotypes up to 4 years prior to the date of analysis) but including in the analysis all animals present in the current official evaluations used as input to MACE (file300Cf). All bulls included in the conventional file300Cr who are also genomically evaluated must be included in the genomic file300Gr. | |
| Line 77: | Line 79: |
| If a significant number of foreign bulls are included in the reference population for national genomic evaluations, and estimations of genomic prediction equations use de-regressed MACE values for these bulls as input, the reduced genomic evaluation can be achieved in three ways, listed by descending order of preference below: | |
| Line 78: | Line 81: |
| ^a^The GEBVtest software (gebvtest.py) uses a trait-independent format (File300). Users can either prepare data in the new format or use the program gtconvert.py to convert the current format into the File300 format.<<BR>> ^b^All other variables should be the same as in the Cxxxf files. | 1. The NGEC can participate in the Interbull truncated-MACE service. By providing reduced-data national EBV to Interbull for truncated MACE, the results returned by Interbull will be the ideal MACE input for reduced-data national genomic evaluation. |
| Line 80: | Line 83: |
| === Specific instructions for data preparation: === a. The domestic bulls (type of proof ≠ 21 or 22) that have EDC≥20 and EDCr = 0 are called '''test bulls.''' Interbull recommends that number of test bulls would be about 0.25 *(number of bulls used as reference population). i. If the number of bulls the country includes in the genomic evaluation is too small, then the accuracy of the GEBVs calculated using the truncated data becomes significantly smaller than with the full data. In that case, the country can use n < 4 years as the time difference between full and reduced data sets. i. If the number of test bulls is too small (ntb < 50), the country may chose to consider foreign bulls (type of proof = 21 or 22) that have EDC≥20 and EDCr = 0 also as test bulls. i. In both exceptions above, the Interbull Centre must be communicated in detail about the criteria adopted to define the test bulls. a. Appropriate time windows (birth years of bulls) may vary depending of the trait to be validated, the speed of their progeny test program and other factors. A shift of the time window with one year will give a different set of bulls that qualify for the test. The standard adopted for the GEBV test is to include four years of candidate/test bulls, which corresponds to an age cutoff of (YYYY-8). For instance, if the Cxxxf is from 2012, Cxxxr and Gxxxr should include performance records up to 2008 and test bulls would be those born from 2004 and 2008. Countries may include more birth years, but the Interbull Centre must be communicated about the reason. a. GEBV is the genomically enhanced breeding value. Correspondingly, the GEDC is a genomically enhanced EDC that combines the EDC from national non-genomic evaluation with the gain from genomic evaluation. This means that GEDC should be larger than EDC and GEDCr should be larger than EDCr. a. Include all the bulls having GEBVr in the data without data edits based on EDC, EDCr, GEDC or GEDCr. a. If GEDCr is not available, then GEDCr = λ * r^2^,,GEBVr,, / (1- r^2^,,GEBVr,,) a. The method of estimation of GEDCr (and/or r^2^,,GEBVr,,) has to be reported in the Interbull GENO form. a. The GEBVr prediction equations also have to be based on the truncated data. If the GEBVr combines information of DGV and EBV (i.e. PA), the EBV (PA) information has to be also from the truncated data. a. Bulls with EBV in the full data sets that have no progeny information four years ago (EDCr=0), should be included in the reduced data set. a. If the EBVs from evaluations published four years ago are available, the country can use these values for the reduced data sets. However, if the evaluation model, trait definitions, etc. have changed from the estimation of EBVs in the reduced data sets and the estimation of EBVs in the full data sets, the GEBVr can be expected to have lower accuracy than GEBV. In this case, the country should report the expected correlation between the old (reduced) and the new (full) data EBVs (see Interbull Testing Method 3). a. In order to remove any change in scale of proof expression, EBVr and GEBVr should be rescaled to the same scale as EBVs, using bulls already proven in the reduced data sets. |
1. The Interbull Centre can make historical files available upon request, which shall include the official MACE results published 4 years earlier. These MACE proofs will be less ideal than truncated MACE proofs, because current evaluation systems were not re-run with older data by any country for the MACE proofs already computed 4 years earlier. 1. The current MACE proofs can be used by excluding all recently proven bulls in MACE who did not have an official MACE proof 4 years earlier. This approach is an exception that should only be used if both preferred options above are impractical. The main concern with this approach is that reduced-data genomic prediction equations will include contributions from phenotypes in most recent 4 years, through sires of the recently proven bulls, and more generally through MACE proofs of all older bull with any relationship to the validation test bulls whose MACE proofs are being excluded. == Specific instructions for data preparation: == A. The domestic bulls (type of proof ≠ 21 or 22) that have EDCf ≥ 20 and EDCr = 0 are called test bulls. Test bulls are likely to be included in the genomic reference population with full data, but not with reduced data. Interbull recommends that the reduction in size of the genomic reference population, due to the dropping of test bulls in reduced data, should not exceed 25% i. If the size of genomic reference population is reduced by too much, then the accuracy of GEBV calculated from truncated data becomes significantly lower than with full data. In that case, the country can use n<4 years as the time difference between full and reduced data sets. i. If the number of test bulls is too small (<50), then the country may choose to also include foreign bulls that have been used locally (type of proof = 21 or 22) with EDCf ≥ 20 local progeny and EDCr = 0 as part of the validation group, to increase the number of test bulls. i. In both exceptions above, the criteria used to define test bulls must be communicated to the Interbull Centre. A. Appropriate time windows (birth years of test bulls) may vary depending on the trait to be validated, the speed of progeny test programs and other factors. The standard adopted for the GEBV test is to include progeny-proven bulls born since (YYYY-8) as test bulls. For instance, if the evaluation year is 2024 and the most recently proven bulls in file300Cf were born in 2020, then the test bulls would include bulls born between 2016 and 2020. Countries may include a wider window of test bulls, or may shift the window by one year, but the reasons must always be communicated to the Interbull Centre. A. Include all available bulls of interest, as described below, in the respective files with their EBVf, EBVr, GEBVf and GEBVr, without editing based on EDCf or EDCr. These final edits, as required for the validation test, are applied within the GEBV test software. A. If the GEBV are a combination of DGV and EBV, then both the DGVr and EBVr used to generate the GEBVr must be estimated from the truncated data. A. Bulls with EBV in the full data sets only, having no progeny information four years ago (EDCr=0), should be included in the reduced-data files (300Cr and 300Gr). Additionally, a minimum of 10 years of bulls with progeny-based EBV in both the full and reduced data sets should be included in the reduced-data files. After recent updates to the software, bulls with progeny in the reduced data are now additionally required as a statistical control group used to improve statistical tests for bias in the evaluations of validation test bulls. |
| Line 96: | Line 104: |
| === Testing for bias === The bias in the national genomic evaluations will be tested using a regression model: |
== Testing for bias in the GEBV == * '''Methodology updates in 2024 ''' |
| Line 99: | Line 107: |
| φ,,i,,= b,,0,, + b,,1,,*GEBVr,,i,, + e,,i,, ''' [1]''' , | The official Interbull GEBV test is now based on !VanRaden's de-regressed GEBV (described in the 2021 Interbull bulletin paper, https://journal.interbull.org/index.php/ib/article/view/82) as the official prediction target. The !VanRaden dGEBV replaces the previously used dEBV target described by Mantysaari et al (2010). Predicting later GEBV or dGEBV from earlier GEBV is conceptually easier to understand and to verify than predictions of dEBV. The new tests are also more suitable for validating single-step models, where genomic preselection effects are properly accounted in GEBVf, GEBVr and dGEBV, whereas dEBV include genomic preselection bias. |
| Line 101: | Line 109: |
| where φ,,i,, is the D_PGM (or DD, if available) from the bulls that have EDC≥20 and EDCr =0. The EDCs from the '''''full data set''''' can be used as weights in the model if DDs are supplied, otherwise the accuracy of the D_PGMs (u,,i,,=EDC/(EDC+λ)) will be used as weights. | With the implementation of a new validation target, the de-regression method has now been internationally standardized because the dGEBV are derived directly from values based on official publication rules, in file300Gf and file300Gr files, in the same way for all countries. |
| Line 103: | Line 111: |
| a. This model is used to estimate b,,1,, to compare with the expectation of b,,1,, (H,,0,,: b,,1,, = E(b,,1,,)) and therefore test the bias on GEBVr. Item 4.3 describes how the expectation of b,,1,, can be derived considering the impact of selective genotyping among test bulls. i. The statistical significance will be tested using a t-test against H,,0,, (C.I. = 0.95). i. For larger populations the estimated standard error might become very small and then the t-test may become too restrictive. In those cases, a “biological significance” will be adopted to test H,,0,, [P((E(b,,1,,)-0.1) ≤ b,,1,, ≤ (P(E(b,,1,,)+0.1))]. i. The country-trait-breeds will pass the test, if b,,1,, value is greater than the lower endpoint of the 95% confidence interval or its biological equivalent. a. The accuracy of GEBVr will be estimated from the R^2^ of the model (accuracy of the model after selection for genotyping). This validation accuracy R^2^,,validation,, = R^2^/ ū, where ū is the average weight of all the test bulls. It will be expected that the mean of published bull r^2^,,GEBVr,, is in agreement with R^2^,,validation,,. ''' ''' ''' ''' |
The software will make sure that full and reduced-data evaluations are on the same genetic base of expression by adjusting the mean and variance of reduced-data evaluations to match the base of expression of full-data evaluations. These adjustments to align the evaluation scales are based on bulls already progeny-proven in the reduced data who have expected changes in evaluations very close or equal to zero, due to either no new progeny or relatively few in the recent data. After aligning the evaluation scales, changes in evaluations for the validation test bulls, who have all their progeny in the recent data, are equivalent to contrasts of evaluation changes for validation bulls relative to previous generations of proven bulls who have expected changes of 0. |
| Line 109: | Line 113: |
| === Testing the improvement from conventional evaluation === The improvement of the added daughter information to the parental information will be estimated by comparing the R^2^ from model [1] with the R^2^ from model [2]: |
Average changes in evaluation between reduced and full data will now have an expectation of 0 for any group of bulls, after the scales are aligned. Additional tests have been added, which account for both the combination of intercept and slope estimates from the validation models, to detect probabilities of bias in below-average versus average versus above-average (top) bulls. A new user option to output base-adjusted evaluations from reduced data, for all or selected traits, can also be used to help isolate reasons for detected biases in the evaluations of any traits failing the GEBV test. |
| Line 112: | Line 115: |
| ''' '''φ,,i,, = b,,0,, + b,,1,,*EBVr,,i,,+ e,,i,, '''[2]''' , | Besides the application of the official GEBV test, the software also allows users to choose different validation targets for further internal research. Below is the list of available options: |
| Line 114: | Line 117: |
| ''' '''where φ,,i,, and the corresponding weight u,,i,, are the same as in model [1]. The R^2^ from model [1] must be higher than the R^2^ from model [2]. | * file300Df_COUBRD (de-regressed EBV, as used previously in the old test) |
| Line 116: | Line 119: |
| === Estimating the effect of selective genotyping on E(b1) === ''' '''The expected value of b,,1 ,,is 1.0 only if the genotyped test bulls are a representative sample of the bulls in the corresponding age classes. The selection based on EBVs before genotyping will reduce the value of b,,1,, and also the value of R^2^ for model [1]. ''' '''The level of selective genotyping can be approximated from the difference between the mean EBV of the genotyped test bulls, µ,,EBVg,,, and the mean EBV of all potential test bulls (i.e. bulls with EDC≥20 and EDC,,r,,=0, genotyped or not), µ,,EBVall,,, and the standard deviation of EBV of all potential test bulls (σ,,EBVall,,). ''' ''' |
* file300Cf_COUBRD (EBV from the full-data evaluation) |
| Line 119: | Line 121: |
| . i = (µ,,EBVg,, - µ,,EBVall,,)/ σ,,EBVall ,,'''[3]'''. ''' ''' | * file300Gf_COUBRD (GEBV from the full-data evaluation) |
| Line 121: | Line 123: |
| Using tables from quantitative genetics books, (e.g. page 379 from Falconer, D. S. & Mackay, T. F. C. ''Introduction to Quantitative Genetics'', Longman, 4^th^ ed. 1996) the proportion of selected (genotyped) individuals (p) can be obtained for the selection differential (i) and the corresponding truncation point x that divides the standard normal density into selected proportion p and non-selected (1-p). ''' '''<<BR>> | * file300Vf_COUBRD (Any user-defined value, e.g. single step DD, new file) |
| Line 123: | Line 125: |
| Having the proportion of the selected individuals, the expected value of the b,,1,, (E(b,,1,,)) and the effect of the selection on R^2^ of the test model can be estimated by approximation of the effect of selection on the variance of the selected trait and on the covariance between the independent (GEBVr) and the dependent (φ) variables. Having (i) as the mean deviation of the selected individuals from the total population in terms of standard deviation from the total population, and (x) as a selection truncation point from the overall mean: | The user must create whichever input file(s) above are needed for the requested validation target. |
| Line 125: | Line 127: |
| k = i(i - x) '''[4]''' | A Bootstrapping approach has been implemented to replace the previous t-test for bias in validation slopes, addressing technical concerns that the t-test was not valid, because validation bulls are genetically related, and the validation model residuals are correlated. |
| Line 127: | Line 129: |
| ''' '''v,,1,, = 1 – k '''[5]''' | The overall validation result, which combines results from either a PASS or FAIL across several sub-tests, will present the following value: PASS, hiSE (i.e. high Standard Error) or FAIL. An overall PASS requires a PASS for the different slope tests plus either a PASS or hiSE for the accuracy test. A result of fail for either the combination of different slope tests or the accuracy test causes an overall FAIL. The new reporting of hiSE indicates too little data to conclusively prove PASS or FAIL in some traits and populations. |
| Line 129: | Line 131: |
| ''' '''Calculating R^2^ before selection (R,,b,,^2^), which is the R^2^ for model [1], from R^2^ after selection | * '''Validation regression models''' |
| Line 131: | Line 133: |
| (R,,a,,^2^): ''' '''R,,b,,^2^ = R,,a,,^2^ / (v,,1,, + kR,,a,,^2^) '''[6]''' | Weighted linear regression models are used to test for bias in both the national genomic and the conventional evaluations, respectively. To pass the official Interbull GEBV test, however, requires only that the GEBVr are unbiased, and not the EBVr. The test for bias in EBVr is provided as comparative and additional information only. |
| Line 133: | Line 135: |
| ''' '''v,,2,, = 1 – kR,,b,,^2^ '''[7]''' | We first define a validation target variable φ that resembles phenotypic progeny averages, and which is based on the progeny contributions in current GEBVf of the validation test bulls. All progeny contributions for the test bulls were from the most recent 4-year period, and contributed to GEBVf and EBVf, but not GEBVr and EBVr. The validation regression models are: |
| Line 135: | Line 137: |
| ''' '''E(b,,1,,) = v,,1,, / v,,2,, '''[8]''' | {{attachment:formula_1_2.png||height="81",width="297"}} |
| Line 137: | Line 139: |
| ''' Example:''' ''' '''Assuming that: µ,,EBVg,, = 16.00; µ,,EBVall,, = 11.76; σ,,EBVall,, = 10.00; R,,a,,^2^ = 0.555. The selection differential (i) for the genotyped bulls equal to 0.424 standard deviations of EBVs (equation [3]), the proportion of genotyped bulls (p) would be 75 percent and the mean deviation of the truncation point from the overall mean (x) would be equals to -0.674 (from reference table). Applying equations [4], [5] and [6] it is possible to calculate R,,b,,^2^ = 0.70. Using equations [7] and [8] E(b,,1,,) = 0.793. | As discussed in the previous section, the validation target φ in the official GEBV test is defined as the dGEBV of !VanRaden (2021). The validation test bulls for both models must meet the following criteria: EDCf ≥ 20 and EDCr = 0, born within a pre-defined range of birth years, such as (YYYY-8) to (YYYY-4) inclusive, where YYYY is the current year of evaluation, and having both an EBVr and a GEBVr available. All validation test bulls with an observation in φ are therefore genotyped, and the most recently progeny proven. |
| Line 139: | Line 141: |
| ''' Table 2 –''' Examples of expected regression coefficients (E(b,,1,,)) as functions of the selection intensity (i) and the coefficient of determination before selection (R,,b,,^2^). ''' '''''' ''' ||<tablewidth="100%" tablestyle="margin-left:.1pt;border-collapse:collapse;border:none;mso-border-alt:solid windowtext .5pt;mso-yfti-tbllook:160;mso-padding-alt:0cm 0cm 0cm 0cm;mso-border-insideh:.5pt solid windowtext;mso-border-insidev:.5pt solid windowtext" tableclass="MsoNormalTable"rowstyle="mso-yfti-irow:0;mso-yfti-firstrow:yes"12% #D9D9D9 height="15.0pt" style="border:solid windowtext 1.0pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm;text-align:center" |2>i ''' ''' ||<12% #D9D9D9 height="15.0pt" style="border:solid windowtext 1.0pt;border-left:none;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt;text-align:center" |2>p ''' ''' ||<12% #D9D9D9 height="15.0pt" style="border:solid windowtext 1.0pt;border-left:none;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt;text-align:center" |2>x ''' ''' ||||||||||<62% #D9D9D9 height="15.0pt" style="border:solid windowtext 1.0pt;border-left:none;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt;text-align:center">E(b,,1,,) ''' ''' || ||<rowstyle="mso-yfti-irow:1"12% #D9D9D9 height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">R,,b,,^2^ = 0.50 ''' ''' ||<12% #D9D9D9 height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">R,,b,,^2^ = 0.55 ''' ''' ||<12% #D9D9D9 height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">R,,b,,^2^ = 0.60 ''' ''' ||<12% #D9D9D9 height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">R,,b,,^2^ = 0.65 ''' ''' ||<12% #D9D9D9 height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">R,,b,,^2^ = 0.70 ''' ''' || ||<rowstyle="mso-yfti-irow:2"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.644 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">60 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">-0.253 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.594 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.619 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.646 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.676 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.709 || ||<rowstyle="mso-yfti-irow:3"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.570 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">65 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">-0.385 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.626 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.650 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.677 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.705 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.736 || ||<rowstyle="mso-yfti-irow:4"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.497 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">70 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">-0.524 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.660 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.683 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.708 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.735 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.764 || ||<rowstyle="mso-yfti-irow:5"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.424 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">75 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">-0.674 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.697 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.718 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.742 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.766 ||<12% #D9D9D9 height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.793 ''' ''' || ||<rowstyle="mso-yfti-irow:6"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.350 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">80 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">-0.842 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.736 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.756 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.777 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.800 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.823 || ||<rowstyle="mso-yfti-irow:7"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.274 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">85 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">-1.036 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.781 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.799 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.817 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.836 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.856 || ||<rowstyle="mso-yfti-irow:8"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.195 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">90 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">-1.282 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.832 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.846 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.861 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.876 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.892 || ||<rowstyle="mso-yfti-irow:9"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.109 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">95 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">-1.645 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.894 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.904 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.914 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.924 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">0.934 || ||<rowstyle="mso-yfti-irow:10;mso-yfti-lastrow:yes"12% height="15.0pt" style="border:solid windowtext 1.0pt;border-top:none;mso-border-top-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:0cm 0cm 0cm 0cm">0.000 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">100 ''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">''' ''' ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">1.000 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">1.000 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">1.000 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">1.000 ||<12% height="15.0pt" style="border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:solid windowtext .5pt;padding:.75pt .75pt 0cm .75pt">1.000 || |
The reliability equivalent of information from progeny phenotypes, all of which were included in the full data but not in the reduced data for the test bulls, is used as the regression weight in both models. The progeny information, expressed as an EDC, is first derived from genomic reliabilities based on full (GRELf) and reduced data sets (GRELr), as shown below, and the EDC are then converted back to a reliability equivalent as the bull's regression weight (WT): |
| Line 152: | Line 143: |
| {{attachment:EDC_formula.png}} | |
| Line 153: | Line 145: |
| The constant λ is a function of the trait heritability but using any value for λ in the pair of equations above will result in the same WT, so these equations can be simplified by substituting λ=1 in both equations. The WT is thus a function of only GRELf and GRELr. | |
| Line 154: | Line 147: |
| * '''Effects of selective genotyping ''' | |
| Line 155: | Line 149: |
| == Interbull form GENO == The methodology for estimation of GEBV and its’ accuracy (r^2^,,GEBV,,) have to be reported by the NGEC in Interbull '''[[https://wiki.interbull.org/public/Form%20GENO?action=print|form GENO]]'''. |
The estimated regression coefficients, b,,1,, and b,,3,, from the two validation models, are compared with expected values to test H,,0,,: b,,1,, = E(b,,1,,) for GEBVr and H,,0,,: b,,3,, = E(b,,3,,) for EBVr. The expected values are equal to 1 for both models if all bulls most recently progeny-proven were also genotyped. The expected values will be lower than 1, however, if only a subset of bulls were genotyped, and the genotyped bulls were non-randomly selected with respect to the given trait. The software includes adjustment for the effects of selective genotyping on E(b,,1,,) and E(b,,3,,). The first step in deriving the adjustments is to estimate selection differentials for each validated trait. Selection differential is the standardized difference in means between genotyped bulls (g) versus (all) progeny-proven bulls who otherwise qualify as members of the validation test group. . i = (µ,,EBVg,, - µ,,EBVall,,)/ σ,,EBVall ,,'''[3]''' Using normal distribution tables from quantitative genetics books (e.g. page 379 from Falconer, D. S. & Mackay, T. F. C. ''Introduction to Quantitative Genetics'', Longman, 4^th^ ed. 1996) the proportion selected by truncation (''p'') to generate an equivalent selection differential as the observed ''i'', and the corresponding truncation point ''x'' that divides the standard normal density into the selected (''p)'' and non-selected (''1-p'') proportions, can be obtained. From the equivalent proportion under truncation selection, the expected values of regression coefficients can be approximated using expected effects of truncation selection on the variances and covariance between φ and the independent variable X, where X is either GEBVr or EBVr. Denoting all variables after selection on φ with a superscript '''s''', defining ''R^2^,,b,,'' = ''R^2^,,(φ,X),,'' before selection, and following Bulmer (1971) and Henderson (1975): {{attachment:formula_4.png||height="168",width="401"}} From the expected ''C^s^(φ,X) and ''V^s^(X) ''after selection, we get the expected b,,1,, after selection: '' ''E^s^(b,,1,,) = ((1 -'' k)'' / ''(''1-''k*R^2^,,b,,)) * ''E(b,,1,,),, ,, '''[5]''' '' ''From the observed R^2^,,φ,x ,, after selection, with the following expected value, we can derive the required ''R^2^,,b,,'' in equation '''[5]''' as follows: '' '' {{attachment:formula_6.png||height="158",width="401"}} '' ''Substituting '''[6]''' into '''[5]''' and simplifying, we get expected slope as a function of observed R^2^,,φ,x ,, '' . '' {{attachment:formula_7.png||height="155",width="536"}} '' '''''Example:''' Let µ,,EBVg,, = 16.00, µ,,EBVall,, = 11.76, σ,,EBVall,, = 10.00, R^2^,,φ,x ,,= 0.50. Using equation [3], the selection differential (''i'') for genotyped bulls equals 0.424. For this value of ''i'', the equivalent proportion by truncation selection for genotyping (''p'') would be 0.75 and the mean deviation of the truncation point from the overall mean (''x'') would be -0.674 (from reference table). From equation [4] we get k=0.466, and from [6] then [5] we get R,,b ,,^2^ = 0.652 and then E^s^ (b,,1,,) = 0.767, or directly from [7] we also get E^s^ (b,,1,,) = 0.767. '' '''''Table 1 -''' Examples of expected regression coefficients (E(b,,1,,)) as functions of the selection intensity (''i'') and the coefficient of determination after selection (R^2^,,φ,x ,,).'' {{attachment:example_expected_regression.png}} * '''Testing for accuracy improvement with genomics ''''' '' The improvement in prediction of daughter performance due to the addition of genomic information (i.e. genotyping) is tested by bootstrapping the difference in validation model R^2^ for models [1] - [2]. A positive difference (P<.05) indicates a significance increase in accuracy with GEBV and therefore results in a Pass. A negative difference (P<.05) results in a Fail, and a non-significant difference (P>.05) indicates that data were insufficient to conclude either way, which therefore results in a designation of hiSE (high standard error). A Pass or hiSE result is required as one part of the overall requirements to PASS the official GEBV test. == Description of National Genomic Evaluations == National Genetic Centres shall provide a description of their national genomic evaluations to Interbull Centre, for now by using the GENO forms, and in the near future, by electronic forms within the PREP database that will be replacing GENO forms. Updated descriptions shall be provided each time changes to the national genomic evaluations are introduced. == References == Sullivan, P.G. 2023. Updated Interbull software for genomic validation tests. Interbull Bulletin 58, p.7-16. !VanRaden, P.M. 2021. Improved genomic validation including extra regressions. Interbull bulletin 56: 65-69. Mäntysaari, E., Liu, Z and !VanRaden P. 2010. Interbull Validation Test for Genomic Evaluations. Interbull Bulletin 41, p. 17-21. Bulmer, M.G. 1971. The effect of selection on genetic variability. American Nat. 105:201. Henderson, C.R. 1975. Best Linear Unbiased estimation and prediction under a selection model. Biometrics 31:423-447. Falconer, D. S. & Mackay, T. F. C. Introduction to Quantitative Genetics, Longman, 4^th^ ed. 1996 |
Interbull CoP - Appendix VIII - Interbull validation test for genomic evaluations - GEBV test
Document based on:
Sullivan, P.G. 2023. Updated Interbull software for genomic validation tests. Interbull Bulletin 58, p.7-16.
VanRaden, P.M. 2021. Improved genomic validation including extra regressions. Interbull bulletin 56: 65-69.
Mäntysaari, E., Liu, Z and VanRaden P. 2010. Interbull Validation Test for Genomic Evaluations. Interbull Bulletin 41, p. 17-21.
Definitions:
- EBV - Estimated Breeding Value (conventional national evaluations of the trait, free of genomic information, which are submitted to Interbull to be used in MACE evaluations)
- DGV - Direct Estimated Genomic Value (genomic evaluations based on SNP prediction equations)
- GEBV - Genomically Enhanced Estimated Breeding Value (evaluations that combine EBV and DGV)
- dGEBV - De-regressed GEBV
- GREL - Genomic reliability of the bull's GEBV
- EDC - Effective Daughter Contribution
- MACE - Multiple Trait Across Country Evaluation
- PA - Parent Average
- DD - Daughter Deviation
- NGEC - National Genetic Evaluation Centre
Motivation
The inclusion of national genomic information in international comparisons for dairy breeds requires that the national genomic breeding values (GEBV) get validated by Interbull in a similar fashion that conventional EBV are validated as a pre-condition to participate in the MACE evaluations.
The GEBV test will be applied to validate national models used to compute GEBV that the national genetic evaluation centers (NGEC) publish and will eventually submit to Interbull for international genetic evaluations including genomic information. The GEBV test can also be considered a quality assurance assessment for national genomic evaluations. GEBV from models that have been tested can be referred to as breeding value estimates with appropriate reliability, and which can be converted to other country scale breeding values using conversion equations derived by Interbull.
Rationale
The GEBV test evaluates:
- the unbiasedness of the genomic evaluations through the evaluation of
- the consistency of the genetic trend captured by GEBV,
- the consistency of bull rankings before versus after having progeny, and
- the consistency of the variation of GEBV relative to EBV;
- the improvement in selection accuracy from the use of GEBV instead of EBV.
A time-oriented cross-validation is used to test how well genomic evaluations of young bull calves, using current models and phenotypic data from 4 years ago, can predict current progeny performance. The NGEC shall re-run their current evaluation software while excluding the most recent 4 years of daughter phenotypes, to obtain reduced-data genetic (EBVr) and genomic (GEBVr) evaluations. The software will then test if the ranking and variance of bull GEBVr match statistical and genetic expectations relative to ranking and variance of the bull comparisons based on current progeny differences, as an indication of unbiasedness. Furthermore, if the GEBVr are more highly correlated than EBVr with the current progeny phenotypes, it is an indication of accuracy improvement with GEBV.
Linear regression models are used for the validation test, where the expected value of regression slopes equals 1 if validation bulls are an unselected group, and a value less than 1 if only a selected subgroup of the most recent proven bulls have been genotyped. The expected slope is lower with selective genotyping due to effects of selection on variances and covariances used to compute the validation slope. The software will account for effects of selective genotyping on expected slopes, using estimates of selection differential from the differences between average EBV of the genotyped bulls versus all proven bulls born in the period considered for validation testing. Bootstrapping is used for all significance testing, and a combination of statistical and biological limits of tolerance is used by Interbull to assign an overall assessment of pass or fail.
Test data sets
Data formats are described at https://interbull.org/ib/gebvtest_software_2024
Full data sets
Two sets of currently official evaluations for progeny-proven bulls shall be provided for the GEBV test. These will be the EBV and GEBV published or otherwise indirectly used by the NGEC for national selection programs. All bulls provided to Interbull in file300 for MACE shall be included in a conventional EBV file (file300Cf) for the GEBV test, and all these same bulls who are genotyped and have a national GEBV shall be included in the GEBV file (file300Gf).
Conventional national genetic evaluation file (file300Cf)
The national EBV sent by the NGEC as input for the most recent Interbull MACE evaluation will be used to identify validation test candidate bulls, estimate the intensity of selective genotyping, and check bulls birth year and type of proof.
Official national genomic evaluation file (file300Gf)
The national GEBV of current MACE bulls will be used to derive target values reflecting unbiased estimates of average progeny performance for the validation test bulls. The official validation target is derived internally by the software, based on the consistent application for all NGEC of a standardized international method for dGEBV developed by VanRaden(2021).
Reduced data sets
The reduced data sets should be prepared by truncating the phenotypes used as input for both the conventional and the genomic evaluations. The NGEC must exclude phenotypic information from the most recent 4 years and re-run the current models of genetic and genomic evaluation for the traits of interest. The pedigree should not be truncated, just the phenotypes, because each validation bull's predicted genetic contributions in future progeny, based solely on the bull's parent average (EBVr=PA) and on PA plus genomic prediction equations (GEBVr) from the reduced-data evaluations will be needed for the validation test.
Reduced conventional genetic evaluation file (file300Cr)
The NGEC shall carry out a conventional genetic evaluation with no genotypes, while using the truncated data (only phenotypes up to 4 years prior to the date of analysis) but including in the analysis all animals present in the current official evaluations used in MACE (file300Cf).
A minimum of 10 most recent birth years of proven bulls included in file300Cf must also be included in file300Cr. The older proven bulls, with progeny proofs already in the reduced data, are required as a comparative control group, to contrast evaluation changes for younger bulls in the validation test group relative to the older control bulls.
Reduced genomic evaluation file (file300Gr)
The NGEC shall carry out a genomic evaluation that includes the genotypes, while using the truncated data (only phenotypes up to 4 years prior to the date of analysis) but including in the analysis all animals present in the current official evaluations used as input to MACE (file300Cf). All bulls included in the conventional file300Cr who are also genomically evaluated must be included in the genomic file300Gr.
If a significant number of foreign bulls are included in the reference population for national genomic evaluations, and estimations of genomic prediction equations use de-regressed MACE values for these bulls as input, the reduced genomic evaluation can be achieved in three ways, listed by descending order of preference below:
- The NGEC can participate in the Interbull truncated-MACE service. By providing reduced-data national EBV to Interbull for truncated MACE, the results returned by Interbull will be the ideal MACE input for reduced-data national genomic evaluation.
- The Interbull Centre can make historical files available upon request, which shall include the official MACE results published 4 years earlier. These MACE proofs will be less ideal than truncated MACE proofs, because current evaluation systems were not re-run with older data by any country for the MACE proofs already computed 4 years earlier.
- The current MACE proofs can be used by excluding all recently proven bulls in MACE who did not have an official MACE proof 4 years earlier. This approach is an exception that should only be used if both preferred options above are impractical. The main concern with this approach is that reduced-data genomic prediction equations will include contributions from phenotypes in most recent 4 years, through sires of the recently proven bulls, and more generally through MACE proofs of all older bull with any relationship to the validation test bulls whose MACE proofs are being excluded.
Specific instructions for data preparation:
- The domestic bulls (type of proof ≠ 21 or 22) that have EDCf ≥ 20 and EDCr = 0 are called test bulls. Test bulls are likely to be included in the genomic reference population with full data, but not with reduced data. Interbull recommends that the reduction in size of the genomic reference population, due to the dropping of test bulls in reduced data, should not exceed 25%
If the size of genomic reference population is reduced by too much, then the accuracy of GEBV calculated from truncated data becomes significantly lower than with full data. In that case, the country can use n<4 years as the time difference between full and reduced data sets.
If the number of test bulls is too small (<50), then the country may choose to also include foreign bulls that have been used locally (type of proof = 21 or 22) with EDCf ≥ 20 local progeny and EDCr = 0 as part of the validation group, to increase the number of test bulls.
- In both exceptions above, the criteria used to define test bulls must be communicated to the Interbull Centre.
- Appropriate time windows (birth years of test bulls) may vary depending on the trait to be validated, the speed of progeny test programs and other factors. The standard adopted for the GEBV test is to include progeny-proven bulls born since (YYYY-8) as test bulls. For instance, if the evaluation year is 2024 and the most recently proven bulls in file300Cf were born in 2020, then the test bulls would include bulls born between 2016 and 2020. Countries may include a wider window of test bulls, or may shift the window by one year, but the reasons must always be communicated to the Interbull Centre.
- Include all available bulls of interest, as described below, in the respective files with their EBVf, EBVr, GEBVf and GEBVr, without editing based on EDCf or EDCr. These final edits, as required for the validation test, are applied within the GEBV test software.
- If the GEBV are a combination of DGV and EBV, then both the DGVr and EBVr used to generate the GEBVr must be estimated from the truncated data.
- Bulls with EBV in the full data sets only, having no progeny information four years ago (EDCr=0), should be included in the reduced-data files (300Cr and 300Gr). Additionally, a minimum of 10 years of bulls with progeny-based EBV in both the full and reduced data sets should be included in the reduced-data files. After recent updates to the software, bulls with progeny in the reduced data are now additionally required as a statistical control group used to improve statistical tests for bias in the evaluations of validation test bulls.
Test description
Testing for bias in the GEBV
Methodology updates in 2024
The official Interbull GEBV test is now based on VanRaden's de-regressed GEBV (described in the 2021 Interbull bulletin paper, https://journal.interbull.org/index.php/ib/article/view/82) as the official prediction target. The VanRaden dGEBV replaces the previously used dEBV target described by Mantysaari et al (2010). Predicting later GEBV or dGEBV from earlier GEBV is conceptually easier to understand and to verify than predictions of dEBV. The new tests are also more suitable for validating single-step models, where genomic preselection effects are properly accounted in GEBVf, GEBVr and dGEBV, whereas dEBV include genomic preselection bias.
With the implementation of a new validation target, the de-regression method has now been internationally standardized because the dGEBV are derived directly from values based on official publication rules, in file300Gf and file300Gr files, in the same way for all countries.
The software will make sure that full and reduced-data evaluations are on the same genetic base of expression by adjusting the mean and variance of reduced-data evaluations to match the base of expression of full-data evaluations. These adjustments to align the evaluation scales are based on bulls already progeny-proven in the reduced data who have expected changes in evaluations very close or equal to zero, due to either no new progeny or relatively few in the recent data. After aligning the evaluation scales, changes in evaluations for the validation test bulls, who have all their progeny in the recent data, are equivalent to contrasts of evaluation changes for validation bulls relative to previous generations of proven bulls who have expected changes of 0.
Average changes in evaluation between reduced and full data will now have an expectation of 0 for any group of bulls, after the scales are aligned. Additional tests have been added, which account for both the combination of intercept and slope estimates from the validation models, to detect probabilities of bias in below-average versus average versus above-average (top) bulls. A new user option to output base-adjusted evaluations from reduced data, for all or selected traits, can also be used to help isolate reasons for detected biases in the evaluations of any traits failing the GEBV test.
Besides the application of the official GEBV test, the software also allows users to choose different validation targets for further internal research. Below is the list of available options:
- file300Df_COUBRD (de-regressed EBV, as used previously in the old test)
- file300Cf_COUBRD (EBV from the full-data evaluation)
- file300Gf_COUBRD (GEBV from the full-data evaluation)
- file300Vf_COUBRD (Any user-defined value, e.g. single step DD, new file)
The user must create whichever input file(s) above are needed for the requested validation target.
A Bootstrapping approach has been implemented to replace the previous t-test for bias in validation slopes, addressing technical concerns that the t-test was not valid, because validation bulls are genetically related, and the validation model residuals are correlated.
The overall validation result, which combines results from either a PASS or FAIL across several sub-tests, will present the following value: PASS, hiSE (i.e. high Standard Error) or FAIL. An overall PASS requires a PASS for the different slope tests plus either a PASS or hiSE for the accuracy test. A result of fail for either the combination of different slope tests or the accuracy test causes an overall FAIL. The new reporting of hiSE indicates too little data to conclusively prove PASS or FAIL in some traits and populations.
Validation regression models
Weighted linear regression models are used to test for bias in both the national genomic and the conventional evaluations, respectively. To pass the official Interbull GEBV test, however, requires only that the GEBVr are unbiased, and not the EBVr. The test for bias in EBVr is provided as comparative and additional information only.
We first define a validation target variable φ that resembles phenotypic progeny averages, and which is based on the progeny contributions in current GEBVf of the validation test bulls. All progeny contributions for the test bulls were from the most recent 4-year period, and contributed to GEBVf and EBVf, but not GEBVr and EBVr. The validation regression models are:
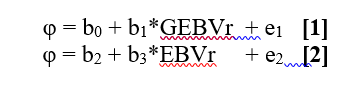
As discussed in the previous section, the validation target φ in the official GEBV test is defined as the dGEBV of VanRaden (2021). The validation test bulls for both models must meet the following criteria: EDCf ≥ 20 and EDCr = 0, born within a pre-defined range of birth years, such as (YYYY-8) to (YYYY-4) inclusive, where YYYY is the current year of evaluation, and having both an EBVr and a GEBVr available. All validation test bulls with an observation in φ are therefore genotyped, and the most recently progeny proven.
The reliability equivalent of information from progeny phenotypes, all of which were included in the full data but not in the reduced data for the test bulls, is used as the regression weight in both models. The progeny information, expressed as an EDC, is first derived from genomic reliabilities based on full (GRELf) and reduced data sets (GRELr), as shown below, and the EDC are then converted back to a reliability equivalent as the bull's regression weight (WT):
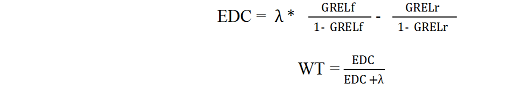
The constant λ is a function of the trait heritability but using any value for λ in the pair of equations above will result in the same WT, so these equations can be simplified by substituting λ=1 in both equations. The WT is thus a function of only GRELf and GRELr.
Effects of selective genotyping
The estimated regression coefficients, b1 and b3 from the two validation models, are compared with expected values to test H0: b1 = E(b1) for GEBVr and H0: b3 = E(b3) for EBVr. The expected values are equal to 1 for both models if all bulls most recently progeny-proven were also genotyped. The expected values will be lower than 1, however, if only a subset of bulls were genotyped, and the genotyped bulls were non-randomly selected with respect to the given trait. The software includes adjustment for the effects of selective genotyping on E(b1) and E(b3).
The first step in deriving the adjustments is to estimate selection differentials for each validated trait. Selection differential is the standardized difference in means between genotyped bulls (g) versus (all) progeny-proven bulls who otherwise qualify as members of the validation test group.
i = (µEBVg - µEBVall)/ σEBVall [3]
Using normal distribution tables from quantitative genetics books (e.g. page 379 from Falconer, D. S. & Mackay, T. F. C. Introduction to Quantitative Genetics, Longman, 4th ed. 1996) the proportion selected by truncation (p) to generate an equivalent selection differential as the observed i, and the corresponding truncation point x that divides the standard normal density into the selected (p) and non-selected (1-p) proportions, can be obtained.
From the equivalent proportion under truncation selection, the expected values of regression coefficients can be approximated using expected effects of truncation selection on the variances and covariance between φ and the independent variable X, where X is either GEBVr or EBVr. Denoting all variables after selection on φ with a superscript s, defining R2b = R2(φ,X) before selection, and following Bulmer (1971) and Henderson (1975):
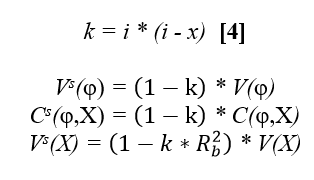
From the expected Cs(φ,X) and Vs(X) after selection, we get the expected b1 after selection:
Es(b1) = ((1 - k) / (1-k*R2b)) * E(b1) [5]
From the observed R2φ,x after selection, with the following expected value, we can derive the required R2b in equation [5] as follows:
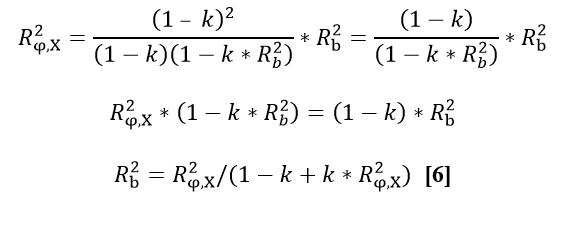
Substituting [6] into [5] and simplifying, we get expected slope as a function of observed R2φ,x
Example: Let µEBVg = 16.00, µEBVall = 11.76, σEBVall = 10.00, R2φ,x = 0.50. Using equation [3], the selection differential (i) for genotyped bulls equals 0.424. For this value of i, the equivalent proportion by truncation selection for genotyping (p) would be 0.75 and the mean deviation of the truncation point from the overall mean (x) would be -0.674 (from reference table). From equation [4] we get k=0.466, and from [6] then [5] we get Rb 2 = 0.652 and then Es (b1) = 0.767, or directly from [7] we also get Es (b1) = 0.767.
Table 1 - Examples of expected regression coefficients (E(b1)) as functions of the selection intensity (i) and the coefficient of determination after selection (R2φ,x ).
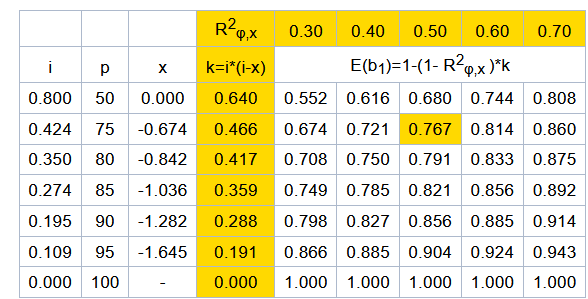
Testing for accuracy improvement with genomics
The improvement in prediction of daughter performance due to the addition of genomic information (i.e. genotyping) is tested by bootstrapping the difference in validation model R2 for models [1] - [2]. A positive difference (P<.05) indicates a significance increase in accuracy with GEBV and therefore results in a Pass. A negative difference (P<.05) results in a Fail, and a non-significant difference (P>.05) indicates that data were insufficient to conclude either way, which therefore results in a designation of hiSE (high standard error). A Pass or hiSE result is required as one part of the overall requirements to PASS the official GEBV test.
Description of National Genomic Evaluations
National Genetic Centres shall provide a description of their national genomic evaluations to Interbull Centre, for now by using the GENO forms, and in the near future, by electronic forms within the PREP database that will be replacing GENO forms.
Updated descriptions shall be provided each time changes to the national genomic evaluations are introduced.
References
Sullivan, P.G. 2023. Updated Interbull software for genomic validation tests. Interbull Bulletin 58, p.7-16.
VanRaden, P.M. 2021. Improved genomic validation including extra regressions. Interbull bulletin 56: 65-69.
Mäntysaari, E., Liu, Z and VanRaden P. 2010. Interbull Validation Test for Genomic Evaluations. Interbull Bulletin 41, p. 17-21.
Bulmer, M.G. 1971. The effect of selection on genetic variability. American Nat. 105:201.
Henderson, C.R. 1975. Best Linear Unbiased estimation and prediction under a selection model. Biometrics 31:423-447.
Falconer, D. S. & Mackay, T. F. C. Introduction to Quantitative Genetics, Longman, 4th ed. 1996