Code of practice for the international genetic/genomic evaluation of dairy bulls at the Interbull Centre¹
¹This document supersedes all aspects of the services provided by the Interbull Centre as described in the original service document of May 25, 1994, titled ”Establishment of a member-funded system for routine international genetic evaluation of dairy bulls at the INTERBULL Centre” and all its subsequent amendments.
1. INTRODUCTION
- The Interbull Centre, located at the Swedish University of Agricultural Sciences (SLU), Uppsala, Sweden, maintains a system for routine computation of international genetic and genomic evaluations of the major groups of dairy breeds and traits. The Interbull Centre receives its responsibilities and authority from the ICAR permanent subcommittee Interbull, as determined by the Letter of Agreement between ICAR and SLU and by the Terms of Reference and Rules of Procedure of the Interbull Sub-Committee. Additionally, the Interbull Centre holds the status of European Union Reference Laboratory for Zootechnics as defined by the Commission decision 96/463/EC, Council decision of the 23rd of July 1996. The services provided by the Interbull Centre aim at facilitating international bull selection and trading within and among the participating countries. Details on the specific services are given in this document. Definitions:
ICAR member – organization that has formally joined the International Committee for Animal Recording (ICAR) and complies with the ICAR statutes and is responsible for the payment of the membership fees.
Interbull subcommittee – permanent subcommittee of ICAR, responsible for coordinating the international genetic evaluation of dairy breeds as well as assisting participating organizations to improve national evaluation systems. The subcommittee is coordinated by a Steering Committee formed by representatives of the participating organizations.
Interbull Centre – section of the Department of Animal Breeding and Genetics of SLU which is the operational unit of Interbull, having received the formal mandate to provide services on behalf of ICAR.
Interbull customer – organization that signs service contracts with the Interbull Centre and is held responsible for the payment of the respective service fees. The Interbull customer may be an ICAR member or act in agreement with the ICAR member representing its country/region.
National genetic evaluation centre (NGEC) – organization that performs regularly the national evaluations and holds the expertise in dairy genetics and breeding.
2. Interbull CoP - Services rendered by the Interbull Centre
2.1 Routine international genetic and genomic evaluations of dairy bulls will be computed for traits and breeds (where applicable) as specified in item 6, and distributed to participating organizations three times per year (item 7.3.1).
2.2 Test evaluation runs will be computed and distributed to participating organizations two times per year (item 7.3.1).
2.3 Coefficients (a- and b-values) for conversions between pairs of participating countries will be computed simultaneously to the international evaluations and distributed to participating organizations. Such coefficients can be used in the interim period between routine international evaluations, provided that no change in base definition has occurred in the participating countries.
2.4 International pedigree files will be collated on the basis of data provided by each participating organization and distributed at each routine and test evaluation runs (item 7.2.8).
2.5 Files with cross-reference information will be extracted from the pedigree database at each routine run and distributed to national evaluation centres together with the international predicted merits.
2.6 Verification of genetic trends by validation of national conventional and genomic EBVs as a pre-requisite for data inclusion in international evaluations.
2.7 For the organizations participating in genomic evaluations, direct genomic value (DGV), polygenic effects, and genomically enhanced breeding values (GEBV) are computed and distributed to participating organizations in all routine and test runs.
2.8 For the organizations participating in the genomic evaluations, imputed genotypes of male animals for each specific participating organization are computed and distributed in all routine and test evaluations.
2.9 Editing and publication of the Interbull Bulletin, containing the proceedings of Interbull annual meetings and technical workshops.
2.10 Maintenance of the Interbull web page and related online services.
2.11 Information about national genetic and genomic evaluation systems, as provided by national members, are collected and made available on the web page of the Interbull Centre.
2.12 Access to information on, and possibilities to discuss, national and international genetic evaluation procedures are provided on the web page of the Interbull Centre and at annually arranged Open Meetings.
3. Interbull CoP - Prerequisites for participation
3.1 Organizations that participate with data must be members of ICAR, and should support the ICAR guidelines for data collection, the principles outlined in “Interbull guidelines for national & international genetic evaluation systems in dairy cattle with focus on production traits” (Interbull Bulletin no. 28) and, where applicable, “Recommended procedures for international use of sire proofs” (Interbull Bulletin no. 4).
3.2 Organizations that participate with data in the MACE evaluations must sign a service contract with the Interbull Centre, and sign letters of agreement covering the trait-groups, in addition to dairy-production traits, that a country participates in the evaluation with data. Likewise, participation in Interbull genomic evaluations must be preceded by the signature of a specific contract with the Interbull Centre. The service contracts and letters of agreement can be found in Appendix I.
3.3 Organizations that participate with data must accept their responsibility as stated in these codes of practice.
3.4 Organizations that do not participate with data, but wish to receive international evaluation results, must be members of ICAR, pay agreed fees (item 10), and sign a service contract and the appropriate letters of agreement with the Interbull Centre.
The process of becoming part of the Interbull Centre services is illustrated graphically in Figure 3.1.
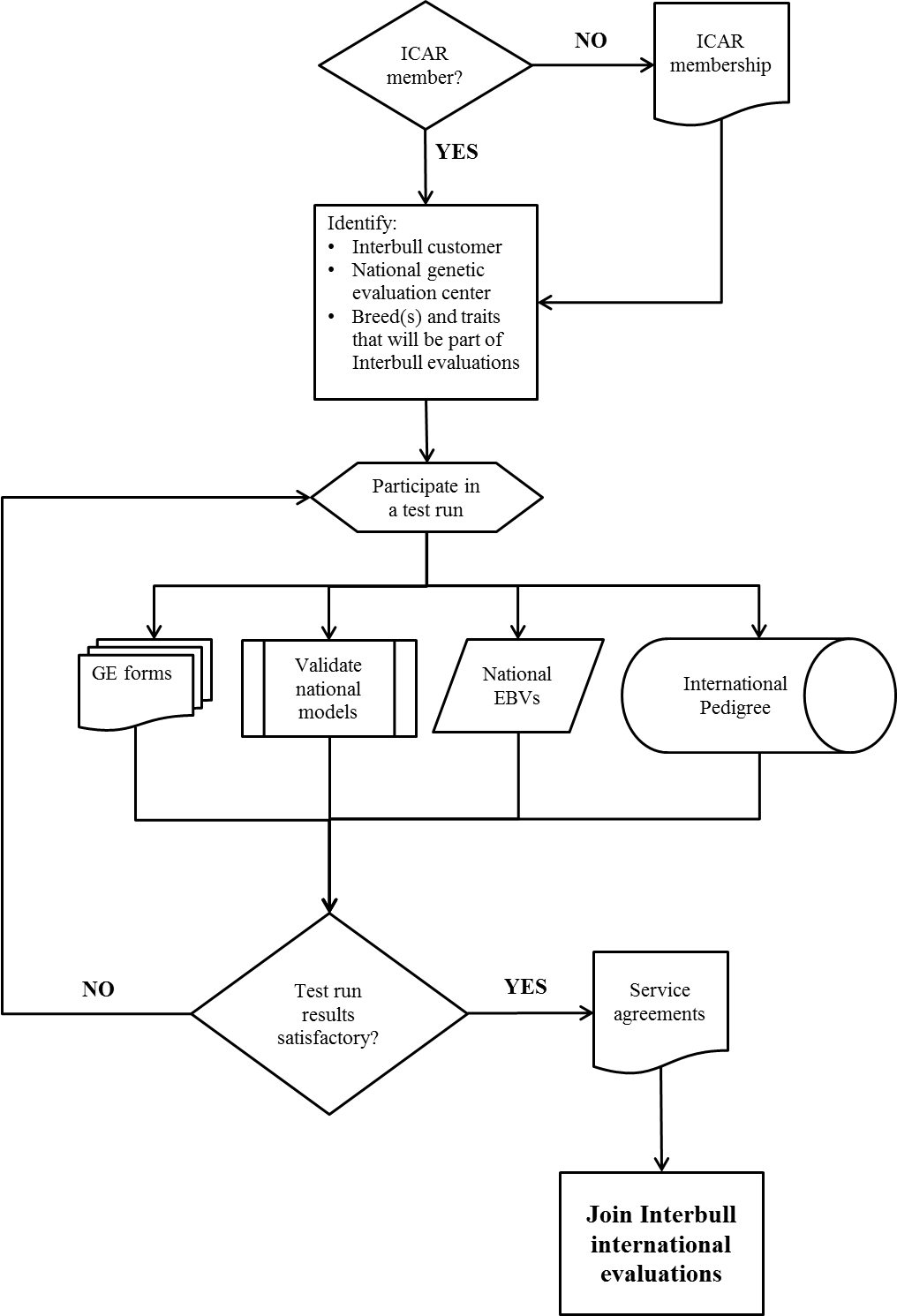
Figure 3.1. Steps for an organization willing to join the Interbull services.
4. Interbull CoP - Responsibilities
All transactions will be carried out between the Interbull Centre and participating organizations.
4.1 Each participating organization will be responsible for:
4.1.1 Checking the quality of national data described below (item 7.1) before sending them to the Interbull Centre, using appropriate protocols including, but not limited to, verification tools provided by the Centre, and communicating explanations for inconsistencies discovered when data is sent. The participating organization is also responsible for correcting (minor) errors in the data, such as file format problems, in cooperation with the Interbull Centre (item 4.2.1).
4.1.2 Submitting data to Interbull for inclusion in a test evaluation run before entering the evaluation scheme for the first time and before major changes in national evaluations (either methodology or logistics, processing and storage of data) are introduced.
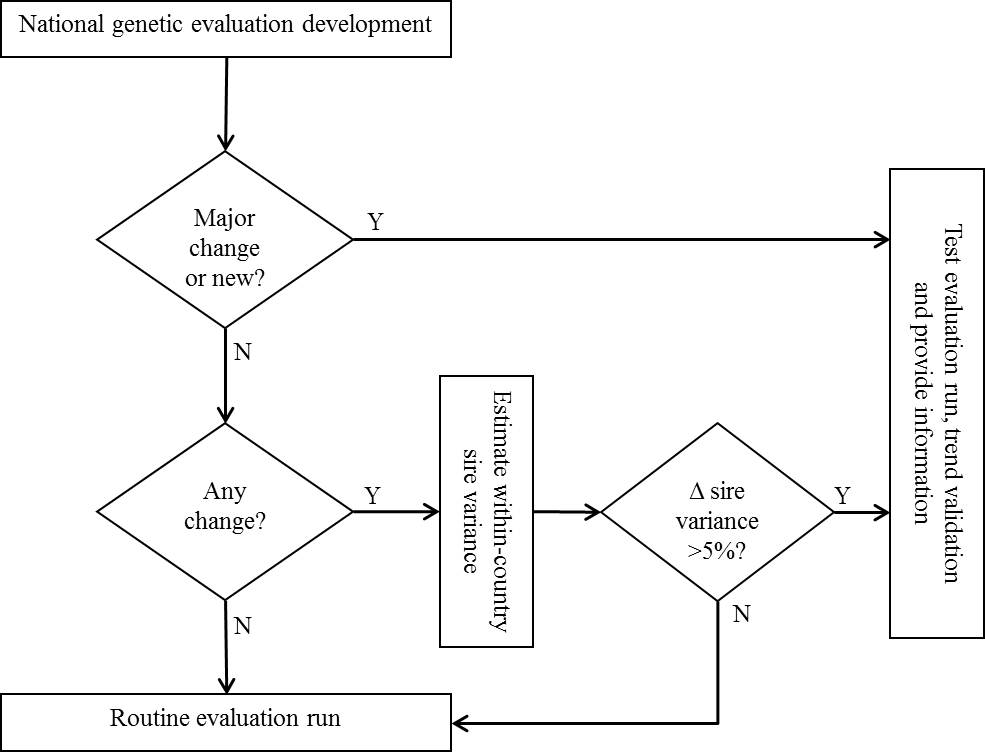
Modified trait definitions, modified models or genetic parameters in breeding value predictions, and modification of pre-adjustment factors, such as for age, are examples of major changes that require participation in a test evaluation run. Modifications of the database, computer programme or compiler, or conversion criterion are examples of changes that may be considered as “minor”, but that may still create “major” differences in results and thus require participation in a test evaluation run. Changes in within‑country sire variance estimates (based on data included in the international evaluation) larger than 5% from one evaluation to the next cannot usually be explained by added information from young bulls, and may be used as an indication that a test evaluation run is required.
The procedural steps are graphically illustrated in Figure 4.1, below.
Partecipation in a test evaluation run with modified data implies that an official implementation of the modification is targeted for a near future, usually within 6 months.
4.1.3 Providing accurate and complete information about national genetic and genomic evaluations using the appropriate forms available in the PREP database (https://prep.interbull.org/) before entering the evaluation scheme for the first time and when changes in national evaluations are introduced.
4.1.4 Validating the estimation of genetic trend and genomic validation test (GEBV test) from the national evaluation, following the procedures in Appendix III and Appendix VIII. Trends should be validated:
a) once before the country enters the national evaluation system for the first time,
b) once every second year,
c) when changes in the national evaluations (methodology or data) are made that require partecipation in a test evaluation run.
Fig. 4.1 Graphical represenation of Interbull actions:
4.1.5 Timely sending national evaluation data files for all actual breeds and traits to the Interbull Centre in the agreed format and media.
4.1.6 Annually providing Interbull with a summary of their publication policy and publication dates. These will be collated by the Interbull Centre and made available to all countries with the understanding that this does not provide permission for publication of results on the scale and base of another country.
4.1.7 Distributing and publishing international evaluation results to appropriate agencies at the national level, upon receiving pertinent files from the Interbull Centre (item 7.2.8).
4.1.8 Access the international bull pedigree file (item 7.2.7) at the Interbull FTP server, carefully examine bulls and pedigree identified with their country code, and solve any dual-registered bulls or discrepancies directly in the IDEA database. It is advised that records for all bulls identified with their country code are provided in the pedigree and national dairy-production file (item 7.1.2, 7.1.3 and 7.1.4) submitted to Interbull for the next evaluation.
4.1.9 Acknowledging the role of the Interbull Centre on the computation of international evaluations whenever these are published and used.
4.1.10 Paying the agreed fees within 30 days after invoices are issued (item 10).
4.1.11 Notifying the Interbull Centre regarding any anticipated future changes in the national evaluation system, including new definition of the reference base, and changes in national identification systems.
4.1.12 Respecting the confidentiality of results from test evaluation runs (item 4.2.4).
4.1.13 Respecting the confidentiality of the pre-release of international evaluation results (item 7.3.1).
4.2 The Interbull Centre is responsible for:
4.2.1 Checking data received from each participating organization and reporting potential errors and/or inconsistencies discovered back to individual organizations; excluding data from the international evaluations if errors or inconsistencies are not corrected before the deadline for receiving data (item 7.3.2) is reached; excluding data from routine international evaluations when data deviate more than expected from previous evaluation, e.g. based on changes in within‑country sire variance estimate (item 4.1.2).
4.2.2 Screening validation results from participating organizations and implementing own validation procedures (Appendix III).
4.2.3 Assessing data connectedness each time a new country joins the scheme.
4.2.4 Conducting test evaluation runs according to the approved schedule (item 7.3.1) investigating major changes in the national evaluation methodology of a country, new breeds and/or traits or new countries entering the scheme, and potential improvements in the international genetic evaluation procedure. Results from test runs will be reviewed by technical representatives of all participating countries prior to incorporation into routine evaluations. Results from test runs are confidential. Genetic evaluation results of individual bulls should not become available outside the official Interbull member representative’s genetic evaluation unit in the participating country. Summary statistics may be shared with relevant advisory groups, in order to prepare for incorporation into routine evaluations, but confidentiality of results must always be maintained and no information may be shared that can be of competitive advantage.
4.2.5 Conducting routine international evaluations according to the approved schedule (item 7.3.1) and the method approved by the Interbull Steering Committee (item 5) utilizing the most recent correct national evaluation results available from the participating organization.
4.2.6 Timely sending results from the international evaluations to participating organizations in the agreed format and media (item 7.2) together with a description of the method and keys to interpretation of the results.
4.2.7 Timely sending results from the international evaluation to countries that do not participate with national data, but meet the prerequisites stated in item 3.
4.3 The responsibilities of the Interbull Steering Committee with reference to the Interbull Centre are to:
4.3.1 Define the general objectives of the Interbull Centre.
4.3.2 Approve new or major modifications of existing services offered by the Interbull Centre upon recommendation from the Interbull Technical Committee.
4.3.3 Approve annual budgets and work plans.
4.3.4 Authorize specific agreements (contracts) between the Interbull Centre and ICAR member and/or participating organizations and/or international societies.
4.3.5 Resolve any problem between the Interbull Centre and a participating organization, or any other collaborating partner, which is not resolved directly by the parties involved.
4.4 The responsibilities of the Interbull Technical Committee are to:
4.4.1 Identify and review technical steps that need to be taken to ensure that an efficient service of high quality is delivered to countries participating in the international genetic evaluations provided by Interbull and to support the continuous development of the service.
4.4.2 Make decisions on methodological issues that may impact evaluation results, but not strategic directions. Examples would include adoption of a proven better algorithm for parameter estimation, or phantom parent group allocation methods.
4.4.3 Make recommendations to the Interbull Steering Committee on methodological issues of such importance that they may affect the service as a whole. Examples would include adding or removing traits from the evaluation, changing editing criteria, or adopting an entirely new method for genetic/genomic evaluation calculation.
4.5 Disclaimer:
Participation in evaluations provided by the Interbull Centre is voluntary and the Interbull Centre and its employees, the Interbull Steering Committee, and/or the Interbull Technical Committee can therefore not be held responsible for financial, or other, damages that may be experienced by anyone utilizing the information contained in reports, listings of international evaluations, and other documents provided as a result of international genetic evaluations. In all events, the maximum liability is equivalent to one annual service fee for the participating organization involved. The Swedish law, applicable to the operations of the Interbull Centre, applies to cases of conflict.
5. Interbull CoP - Methods of international evaluation
5.1. Multiple-trait Across Country Evaluation (MACE)
5.1.1. International evaluations will be computed by a linear multiple-trait across country (MACE) model analysis of national evaluation results from the participating countries.
The model to be used is: Y = c + g + s + e
- Where: Y = De-regressed national genetic evaluations c = Country of evaluation effect g = Genetic group of bull effect, defined by the bull’s population of origin and year of birth s = Bull genetic effect including genetic relationships among bulls in all participating countries e = Residual effect. The international predicted genetic merits will be formed by the sum of the solution for the bull, the genetic group and country effects.
5.1.2. International evaluations will be computed for one trait at a time, and within breed. For breed definition, see item 7.1.7.
5.1.3. Data on all bulls evaluated in each country will be considered in the international genetic evaluation, subject to the following editing criteria:
- only artificial insemination (AI) bulls are included; "other" bulls (status of bull = 20) are considered only if the predicted genetic merit is identified as based on second crop daughters only (type of genetic merit = 21) or imported daughters/embryos (type of genetic merit = 22; see file format for definitions of status of bull and type of predicted genetic merit),
- only bulls with daughters recorded in at least 10 herds are included,
- only bulls born since 1986 and 1981 for Holstein and other breeds, respectively, are included,
- bulls with second country evaluations (type of predicted genetic merit = 21) are included only when the predicted genetic merit meets national standards for official publication in the country of first evaluation of the animal (official publication of predicted genetic merit = Y and type of predicted genetic merit = 11 or 12) OR if the predicted genetic merit is based on minimum 150/30/80 daughters in 50/10/20 herds (Holstein/Guernsey/other breeds) whether or not first country evaluation (type of predicted genetic merit = 11 or 12) is included in the data. Second country data may be excluded in view of scientific evidence of bias, examined by the Interbull Centre
- bulls with second country evaluations based on more than 50% imported daughters/embryos (type of genetic merit = 22) are included only when the predicted genetic merit meets national standards for official publication in the country sending the information (official publication of predicted genetic merit = Y), and first country evaluation (type of predicted genetic merit = 11 or 12) is included in the data.
Exceptions from these requirements may be accepted as part of Interbull standard procedures for a trait-group and specified in the appropriate section of Chapter 6 “Traits and breeds”.
5.1.4. Data on all bulls evaluated in each country will be considered in estimation of genetic correlations between countries, subject to the following editing criteria:
- only artificial insemination (AI) bulls are included; “other” bulls (status of bull = 20) are considered only if the predicted genetic merit is identified as based on second country evaluation (type of predicted genetic merit = 21; see file format for definitions of status of bull and type of predicted genetic merit).
Exceptions from these requirements may be accepted as part of Interbull standard procedures for a trait-group and specified in the appropriate section of Chapter 6 “Traits and breeds”.
5.1.5. Dependent variables in the evaluation model are de-regressed national genetic evaluations.
5.1.6. Variation in the precision of the national predicted genetic merits is accounted for in the linear model by applying a weighting factor (effective daughter contribution, EDC) that considers amount of information, contemporary group structure, correlations of repeated observations within the same animal, and the reliability of daughters’ female ancestors. The calculation of EDC’s has to be implemented separately by each individual organization participating in Interbull evaluations, following the procedure in Appendix IV and V.
5.1.7. Within‑country sire variances are to be estimated during each evaluation run. Correlations between countries are estimated at every test run for all breeds and traits. Any change in national genetic evaluations should be tested in a test run as well as new countries/traits entering the system.
5.1.8. Conversion coefficients among all participating countries and for all traits are computed based on international predicted genetic merits of bull’s that are progeny tested only in one country (country of origin), with minimum birth year 1993 and 1992 for Holstein and other breeds, respectively in year 2004 (to be updated by 1 year at each January evaluation), and a predicted genetic merit based on a minimum 20 herds and with 75% international reliability/repeatability. A minimum of 20 bulls is required to compute such conversions. For country combinations not fulfilling these requirements conversion equations are computed based on theoretical b-values and empirical a-values. The theoretical b-value is computed as rG(i,j)×std(i)/std(j), where rG(i,j) is the genetic correlation between country i and j, and std(j) is the sire standard deviation for country j. The a-value is estimated as m(i)-b×m(j) where m(i) is the mean international proof for country i based on all bulls progeny tested only in one country born since 1996 (Holstein; 1995 for other breeds).
Exceptions from these requirements may be accepted as part of Interbull standard procedures for a trait-group and specified in the appropriate section of Chapter 6 “Traits and breeds”.
5.1.9. Some details on the genetic evaluation procedure are provided in the service documentation made available at http://www.interbull.org and together with each distribution of results from the international evaluations
5.1.10. The international genetic evaluation procedure is based on international work described in the following scientific publications:
- International genetic evaluation computation:
- Schaeffer, 1994. J. Dairy Sci. 77:2671-2678
- Klei, 1998. Interbull Bulletin 17:3-7
- Weighting factors:
- Fikse and Banos, 2001. J. Dairy Sci. 84:1759-1767
- De-regression:
- Sigurdsson and Banos, 1995. Acta Agric. Scand. 45:207-219
- Jairath et al., 1998. J. Dairy Sci. Vol. 81:550-562
- Genetic parameter estimation:
- Sigurdsson et al., 1996. Acta Agric. Scand. 46:129-136
- Klei and Weigel, 1998. Interbull Bulletin 17: 8-14
- Sullivan, 1999. Interbull Bulletin 22:146-148
- Jorjani et al., 2003. J. Dairy Sci. 86:677-679
- Time edits:
- Weigel and Banos, 1997. J. Dairy Sci. 80:3425-3430 5.11
- International reliability estimation:
- Harris and Johnson, 1998. Interbull Bulletin 17:31-36
5.1.11. Newer developments regarding international evaluation methodology will be applied whenever research efforts are completed and considered appropriate, and approved by the Interbull Steering Committee.
5.2. International Genomic Evaluation of Young Bulls (GMACE)
5.2.1. International GMACE evaluations will be computed for one trait at a time, and within breed. For breed definition, see item 7.1.7.
5.2.2. Data on all bulls evaluated in each country will be considered in the GMACE international genetic evaluation, subject to the following editing criteria:
- National GEBV’s will be required to be from the same model and on the same base and scale as the national EBVs of progeny-tested bulls provided for classical MACE. A country can therefore only participate with genomic data in GMACE for the same traits as they participate with in classical MACE.
5.2.3. National GEBV data are edited according to the following criteria:
- Bulls included in the GMACE breeding value predictions:
- no conventional proof and maximum seven years of age
- sire and dam with conventional proof in corresponding conventional evaluation or with GMACE evaluation (e.g. young genomic sire of young genomic bull)
- pedigree in the Interbull pedigree data base
- Genotypes data of bulls participating in international genomic evaluation are not subjected to edits that are based on the number of daughters/herds/EDC.
5.2.4. Dependent variables in the evaluation model are Mendelian Sampling deviations computed as MS = national GEBV – MACE PA.
5.2.5. Correlations between countries are estimated only in classical MACE but utilized also in GMACE.
5.2.6. The Parameter-space approach (Sullivan, 2016) is used in GMACE. Genomic variances are not estimated (the ratio of genomic SD to MACE SD is assumed to be equal to one).
5.2.7. Some details on the genetic evaluation procedure are provided in the service documentation made available at http://www.interbull.org and together with each distribution of results from the international evaluations
5.2.8. The GMACE procedure is based on international work described in the following scientific publications:
- GMACE implementation:
- Sullivan, P.G. 2016. Defining a Parameter Space for GMACE. Interbull Bulletin 50:85-93.
Sullivan, P.G. and VanRaden, P.M. 2010. Interbull Bulletin 41:3-7
- Sullivan, P.G. et al., 2011. Interbull Bulletin 44: 87-94
- Sullivan, P.G. and Jakobsen, J.H. 2012. Interbull Bulletin 45: 3-7.
VanRaden, P.M. and Sullivan, P.G. 2010. Gen. Sel. Evol. 42: 7
- Sullivan, P.G. 2013. GMACE reliability approximation. Interbull Bulletin 47: 1-4
- Sullivan, P.G. 2013. GMACE variance estimation. Interbull Bulletin 47: 5-9
- Sullivan, P.G. 2013. GMACE weighting factors. Interbull Bulletin 47: 10-14.
Sullivan, P.G. & Jacobsen, J.H. 2014. GMACE pilot #4: Adjusting the national reliability input data. Interbull Bulletin 48: 40-45.
Sullivan, P.G. & Jacobsen, J.H. 2014. GMACE without variance estimation. Interbull Bulletin 48: 46-49.
- Validation of national genomic evaluations:
Mäntysaari, E., Liu, Z and VanRaden P. 2011. Interbull Bulletin 41, p. 17-21.
5.3. International Genotype-based Evaluation (InterGenomics)
5.3.1 International genomic evaluations will be computed by an iterative, nonlinear model with heavy-tailed prior for marker effects analogous to Bayes A (a curve parameter of 1.05 is used).
5.3.2 Base population allele frequencies are subtracted from genotypes, and a polygenic effect (poly) additive variance, which value is depending on the trait, is fit in the model.
5.3.3 The model applied is DPGM = mean + Σgenotypes*effects + poly + error.
5.3.4 The InterGenomics procedure is based on international work described in the following scientific publications:
VanRaden, P. (2008) Efficient methods to compute genomic predictions. Journal of dairy Science 91. 4114-4123
VanRaden, P. M. 2011. findhap.f90. Accessed May 25, 2011. http://aipl.arsusda.gov/software/findhap/
6. Interbull CoP - Traits and breeds
6.1 The service considers Brown Swiss, Guernsey, Holstein-Friesian (Black & White, and Red & White), Jersey, Red Dairy Cattle, and Simmental (including Montbeliarde) type bulls (item 7.1.7).
6.2 Dairy-production traits considered are milk, fat and protein yields. Acceptable units of measurement are kilogram, liter, pound, and relative breeding value.
6.3 Conformation traits considered for Guernsey, Holstein-Friesian, Jersey, and Red Dairy Cattle breeds are:
Trait |
Trait definition |
Alternative trait definition |
1. Stature |
height of the rump between hips |
wither height |
2. Chest Width |
width of the fore end of the cow; distance between the fore legs; front view |
strength |
3. Body Depth |
depth of the last rib, deepest point of the body; side view; two dimensional |
depth of chest |
4. Angularity |
angularity is not a single linear trait and cannot be measured; it is an essential trait to describe the perceived milk production; the extreme is a cow that shows openness and angle of the ribs, flatness of bone, sharp shouldered, long neck, clean head and lack of excess fleshing on hips and pins |
dairy character; dairy form |
5. Rump Angle |
angle of the rump from hips to pins; side view |
|
6. Rump Width |
distance between pins |
distance between hips/thurls |
7. Rear Leg set |
angle of the hock in the rear leg; side view |
|
8. Rear Leg Rear View |
angle of toe out of the rear foot |
locomotion |
9. Foot Angle |
angle between ground and front of the rear foot; side view |
foot diagonal; heel depth |
10. Fore Udder |
strength and quality of attachment between body and fore udder; side view |
|
11. Rear Udder Height |
distance between vulva and milk secreting tissue; in relation to the height of the animal |
distance between pin bone and milk secreting tissue |
12. Udder Support |
cleft of udder; emphasis to the bottom of udder |
central ligament; median suspensory |
13. Udder Depth |
distance between hock and bottom of udder |
|
14. Teat Placement |
placement of front teats; rear view |
|
15. Teat Length |
length of front teats; side view |
|
16. Rear Teat Placement |
position of the rear teat from the center of the quarter |
|
17. Overall conformation score |
|
final score; final class |
18. Overall udder score |
|
|
19. Overall feet & leg score |
|
|
20. Body Condition Score |
|
|
21. Locomotion |
|
|
Conformation traits considered for the Brown Swiss breed are:
Trait |
Trait definition |
1. Stature |
|
2. Chest Width |
|
3. Body Depth |
|
4. Angularity |
|
5. Rump Angle |
|
6. Rump Width (Thurl Width) |
|
7. Rear Leg Side View |
|
8. Pasterns/Foot Angle |
|
9. Deep Heel (Hoof Height) |
|
10. Fore Udder Attachment |
|
11. Rear Udder Attachment Height |
|
12. Rear Udder Attachment Width |
|
13. Udder Support |
|
14. Udder Depth |
|
15. Teat Placement |
|
16. Teat Length |
|
17. Rear Teat Placement |
position of the rear teat from the center of the quarter |
18. Overall conformation score |
|
19. Overall udder score |
|
20. Overall feet & leg score |
|
21. Overall Frame |
|
22.Top Line |
|
23 Overall Rump |
|
24 Rump Length |
|
25 Thurl Position |
|
26 Hock Quality |
|
27 Fore Udder length |
|
28 Udder Balance |
|
29 Teat Direction |
|
30 Teat Thickness |
|
6.4 Udder health traits considered are milk somatic cells and mastitis.
6.4.1 For each breed, two separate international genetic evaluations are computed. The first includes milk somatic cell national evaluations from individual countries. The second includes national evaluations for clinical mastitis as a direct trait from countries that make this information available and milk somatic cell national evaluations for all other countries.
6.4.2 For clinical mastitis, bulls are required to have at least 50 daughters in at least 10 herds for inclusion in the international genetic evaluation and estimation of genetic correlations between countries, in addition to the criteria mentioned in items 5.1.3 and 5.1.4.
6.5 Longevity trait considered is direct longevity. National genetic evaluations for direct longevity should be provided if available, even if they are not officially provided in the country, which is an exception from item 7.1.5.
6.6 Calving traits considered are direct and maternal calving ease and direct and maternal stillbirth.
6.6.1 Bulls are required to have at least 50 calves in at least 10 herds for direct traits and at least 50 daughters in at least 10 herds for maternal traits for inclusion in the international genetic evaluation and estimation of genetic correlations between countries, in addition to the criteria mentioned in items 5.1.3 and 5.1.4.
6.7 Female fertility traits considered are maiden heifers ability to conceive, lactating cows ability to recycle after calving, lactating cows ability to conceive measured as a rate trait, lactating cows ability to conceive measured as an interval trait, and lactating cows measurement of interval calving conception.
6.8 Workability traits considered are milking speed and temperament.
6.9 Snp training for clinical mastitis, trait considered is clinical mastitis.
6.9.1 Data shall comply to the following requirements: Number of daughters and herds shall be based ONLY on clinical mastitis records, when genetic reliabilities are estimated with a multiple trait model the corresponding EDC calculation shall not be based on a single trait model.
6.9.2 Country willing to participate for the first time to the Snp training evaluation for clinical mastitis shall provide the following information to the Centre prior of sending their data: average count of cma daughters vs. scs daughters, average count of cma EDC vs. scs EDC, information on method used to calculate cma EDC.
6.10 A description of the national genetic/genomic evaluation system should be provided at each test run for all traits, whose genetic model has been changed, by using the relative electronic forms available in the PREP database (https://prep.interbull.org). Also, the zip file produced by the TestTrend and GEBV test softwares should be provided (before the end of each test run with results pertaining to conventional validation or when available with results pertaining to genomic validation). Access to the TrendTest software is available through IDEA database, access to the GEBV test software is available on the Interbull website www.interbull.org. For more details on Interbull validation methodology please refer to APPENDIX III and VIII
6.11 Conversion coefficients are computed based on international predicted merit of bull’s that are progeny tested only in one country (country of origin), with minimum birth year 1989 in year 2004 (to be updated by 1 year at each January evaluation), in addition to the criteria mentioned in item 5.1.8. For country combinations not fulfilling these requirements, conversion equations are computed based on theoretical b-values and empirical a-values as described in item 5.1.8.
6.12 Consideration of application of international genetic/genomic evaluations on other breeds and other traits will be subject to results of pertinent research and development work. Such work would normally follow the subsequent steps:
6.12.1 Scientific research should be conducted at the Interbull Centre or at other institutes worldwide. Collaborative research proposals with Interbull are scientifically reviewed before they are adopted. Research results are usually presented at the annual Interbull meetings and reviewed by technical representatives of members and by the Interbull Technical Committee.
6.12.2 When such research has advanced, pilot studies are conducted and results are distributed to participating organizations for review.
6.12.3 Results from the pilot studies are reviewed, usually at a technical workshop, with principal investigators and scientists from all member countries.
6.12.4 Pending the outcome of the review of pilot study results, the Interbull Steering Committee decides to proceed with the expansion of the service. A service document is prepared and distributed, and a test evaluation run is scheduled.
6.12.5 Following successful completion and review of the test evaluation run, the new traits/breeds are included in the routine evaluation.
6.13 Deviations from the procedure outlined in item 6.12 may be accepted by the Interbull Steering Committee, for instance when breeds and traits are added to an already established service.
7. Interbull CoP - Data Exchange and time of evaluation
7.1 Data from participating organizations to the Interbull Centre:
7.1.1 Each participating organization is expected to provide:
- A pedigree in a sire-dam format to be uploaded into the IDEA (Interbull Data Exchange Area) database,
- Predicted genetic and genomic merit file for any trait the country participates with data in the international evaluations, and
- File with parameters to be used in the international genetic evaluation.
- All above files must be checked by the relevant checking program available inside IDEA
- The .zip file produced for the pedigree and/or for the predicted geneti/genomic merit plus parameter needs to be uploaded into IDEA
Genotypes to be uploaded to the GenoEx database.
Countries are also expected to prepare files for genetic and genomic validation whenever (major) changes in national evaluations require that data is submitted for a test evaluation run, or when the previous genetic validation was done more than two years ago. Files should be prepared according to the applicable formats available in the Interbull webpage for conventional validation https://interbull.org/ib/validation and/or genomic validation https://interbull.org/ib/gebvtest
200 = pedigree file to be uploaded in IDEA;
300 = bull national predicted genetic merit file (all traits);
301 = parameter to be used in the international genetic evaluation (all traits)
Validation files:
302 = data file for conventional trendtest method 2
303 = data file for conventional trendtest method 3
300Cf = data file containing full conventional genetic evaluation for GEBV test
300Cr = data file containing reduced conventional genetic evaluation for GEBV test
300Gf = data file containing full genomic evaluation for GEBV test
300Gr = data file containing reduced genomic evaluation for GEBV test
Conventional validation files, 302 and 303, should be used to run the conventionalTrendTest software, genomic validation files (Cf, Gf, Cr and Gr) should be used to run the GEBV test software. The zip file produced by the softwares should be sent back to Interbull Centre before the end of each test run evaluation.
7.1.2 Only pedigree information as present in the IDEA database is used for the international evaluation. After the deadline for submission of data has passed the uploading functionality in IDEA is blocked by Interbull Centre staff and the whole IDEA pedigree content is extracted. Breed-specific pedigree files, in a sire-dam format, are then reconstructed starting from all animals having a valid predicted genetic merit for any submitted traits.
7.1.5 Data for the international evaluations will normally be individual trait national predicted genetic/genomic merits (estimated breeding value, transmitting ability, or relative breeding value (RBV)) or SNP effects of bulls in the country’s most recent national evaluation. The predicted genetic/genomic merits should be provided in the same way as it is officially, and publicly, defined and expressed (reference base and official unit of measurement). All traits submitted to Interbull Centre for international evaluations must be described in detail using the PREP database.
Exceptions from these requirements may be accepted as part of Interbull standard procedures for a trait-group and specified in the appropriate section of Chapter 6 “Traits and breeds”.
7.1.6 Countries submitting their data in RBV are particularly requested to include 2 decimal points in the variables, as explained in the file formats. De-regression and international evaluation will consider data in the original unit of measurement, i.e. not as RBV’s, but, as part of the service, international predicted genetic merits will be converted to RBV prior to release.
Countries submitting their data in RBV should provide separately the formulae and coefficients for back-transformation to estimated breeding values or transmitting abilities. Coefficients should be provided each time they are updated and reported in the relevant electronic form within the PREP database.
7.1.7 Bull records from the following breed groups can be sent to the Interbull Centre: Brown Swiss-type, Guernsey-type, Holstein-Friesian-type, Jersey-type, Milking Shorthorn-type, Pinzgauer-type, Red Dairy Cattle-type (including several Red-and-White breeds), Simmental (including Montbeliarde)-type, and non-Red Dairy Cattle Red or Red-and-White-type breeds (e.g. MRY). Other breed groups will be added in the future if needed. Bulls should be classified under one of the above breed groups according to the definition given in each country, e.g. Red Holsteins could be categorized as Holstein-Friesian (if evaluated together with Black & White in a country) or non-Red Dairy Cattle Red depending on the direction the population has taken in this country. Individual countries should identify the breed groups their populations belong to. In case of cross-breeding, the breed with the highest percentage should be considered. International evaluations will be computed within the Brown Swiss, Guernsey, Holstein-Friesian, Jersey, Red Dairy Cattle, and Simmental breed groups, while other breed groups may be added in the future if needed and if investigations have shown the feasibility of corresponding international evaluations.
7.1.8 Data on all bulls (domestic and imported) evaluated in the participating country with daughters in at least 10 herds should be sent to the Interbull Centre. Individual countries are responsible for identifying the records properly with respect to type of predicted genetic/genomic merit, status of predicted genetic/genomic merit, status of bull, etc, as identified in the file format descriptions.
7.1.9 Data from each country are expected to be “clean” from foreign daughter information. Participants are required to address that situation in their submitted files.
7.1.10 Organizations are required to upload data (predicted genetic/genomic merit and parameter) into IDEA database (username and password are provided separately to organizations participating in the international evaluation). Genotypes need to be uploaded to the GenoEx database.
7.1.11 Additional information like GENO Forms should be uploaded into Interbull Centre FTP server (username and password are provided separately to organizations participating in the international evaluation) while percentage of red should be uploaded into IDEA using the AnimInfo module. Additional information pertaining to conventional evaluations should be reported within the PREP database (username and password are provided separately to organizations participating in the international evaluation).
7.1.12 A detailed definition of the reference (genetic) base considered in the national genetic evaluation, as well as description of the base for age adjustment (if applicable), needs to be entered in the relative electronic form available in the PREP database.
7.2 Data from the Interbull Centre to participating organizations:
7.2.1 One international predicted genetic merit file for each trait-group including bulls from all participating countries will be made available in each organization's own site on the Interbull Center FTP server together with information on the file format. The files will include bulls that are:
- identified as officially publishable in at least one country;
- bulls identified as part of a simultaneous progeny-testing program in at least 2 countries. International predicted genetic merits would be as defined in each country (estimated breeding value, transmitting ability, or relative breeding value) expressed in the relevant base and unit.
File formats are:
- For Conventional MACE and GMACE:
030 = Interbull production predicted genetic merit file;
035 = Interbull conformation predicted genetic merit file;
135 = Interbull Brown Swiss conformation predicted genetic merit file;
036 =Interbull milk somatic cell and clinical mastitis predicted genetic merit file;
037 = Interbull longevity predicted genetic merit file;
038 = Interbull calving ease and stillbirth predicted genetic merit file;
039 = Interbull female fertility predicted genetic merit file.
040 = Interbull workability predicted genetic merit file.
041 = Interbull SNP Training for Clinical Mastitis File
For InterGenomics
InterGenomics files have no file code.
7.2.2 One file with international parent average reliability (ipr-files) and predicted parent averages for each trait-group including bulls from all participating organizations will be made available in each organization's own site on the Interbull FTP server together with information on the file format.
File formats are:
060 = Interbull production parent average reliability file;
065 = Interbull conformation parent average reliability file;
165 = Interbull Brown Swiss conformation parent average reliability file;
066 = Interbull milk somatic cell and clinical mastitis parent average reliability file;
067 = Interbull longevity parent average reliability file;
068 = Interbull calving ease and stillbirth parent average reliability file;
069 = Interbull female fertility parent average reliability file.
070 = Interbull workability parent average reliability file.
071 = Interbull SNP Training for Clinical Mastitis File
7.2.4 Each country providing data will receive evaluations for all breeds expressed on their base and scale.
7.2.5 Each country providing data will also receive evaluations for all bulls expressed on the base and scale of all other countries providing data. This information is not to be published by the country receiving it and may only be used for research and internal purposes.
7.2.6 A two-way table with conversion coefficients among all participating countries for all trait-groups evaluated, but subject to limitations identified in item 5.8, will be distributed.
7.2.7 Countries that do not participate with data, but wish to receive results from the international evaluation, will receive the same information as countries providing data. This information is not to be published by the country receiving it. It can only be used for research and internal purposes.
7.2.8 All participating organizations will have access to international pedigree files including all individual bulls. This information will be made available at the Interbull FTP server after each routine evaluation as one file per breed. The information in the pedigree file is strictly confidential and should be handled accordingly, and should only be used to prevent and to resolve identity problems.
7.2.9 Individual countries will be responsible for publishing results of the international evaluations according to own national requirements; publications should distinguish between national predicted genetic merits, international predicted genetic merits and national predicted genetic merits converted using the Interbull conversion factors.
All countries are expected to honor the Interbull “Guidelines for minimum requirements for advertising genetic merit of dairy animals” (Appendix V), and to enforce conformance with the Guidelines by all appropriate agencies at the national and international level.
7.3 Time of evaluations and deadlines:
7.3.1 Routine conventional and genomic international evaluations will be computed three times per year, in April, August and December. Test evaluation runs will be computed three times per year, in January, May and September.
7.3.2 Participating organizations should send their data for MACE routine evaluations to the IDEA database no later than by the Monday 22 days prior to a scheduled release of results. Submission of data for GMACE routine evaluations to IDEA database should be no later than by the Monday 15 days prior to a scheduled release of results. The definitive deadline for sending data is at 17:00 CET, but countries are encouraged to send data at earliest possible opportunity, since it would allow more time for checking and verification of the data, and communicating about possible inconsistencies. The most recent, correct, national evaluation file received by the deadline will be used in the international evaluations.
7.3.3 The target time for release of results for official publication is on the first Tuesday in April, the second Tuesday in August and the first Tuesday in December. The earliest possible official publication of results is at 07:00 local time, on the scheduled Tuesday. Target time for a confidential pre‑release of MACE evaluation results to countries having signed the “Interbull evaluation pre‑release agreement” (Appendix VI) is on the Monday, 12:00 Central European Time (CET), 15 days prior to the scheduled release. Target time for a confidential pre‑release of GMACE evaluation results to countries having signed the “Interbull evaluation pre‑release agreement” (Appendix VI) is on the Tuesday, 12:00 Central European Time (CET), 7 days prior to the scheduled release. The pre‑release of evaluation results is strictly confidential and the sole intent is for checking of results, but it also provides the opportunity for countries to pre‑process the data prior to official release. The Interbull Centre may decide to release new evaluation results if a re-run is necessary, due to errors in procedures at the Centre or in national data discovered by the Centre or by a member after the pre-release. The Interbull Centre should send a report to all members on the likelihood that the Tuesday morning release will be possible to keep, no later than Friday, 17:00 CET.
7.3.4 Data submitted for test evaluation runs (items 4.1.2 and 4.2.4) should be sent by each new country, and by each country introducing changes in national evaluations, according to the following general schedule:
MACE TEST RUN: no later than 17:00 CET the Tuesday three (3) weeks after the release of the April and the August routine runs and six (6) weeks after the release of December routine run. MACE test run results will be made available to countries four (4) weeks after.
GMACE TEST RUN: GEBV test results should be put on the Interbull FTP server no later than 17:00 CET the Tuesday two (2) weeks after MACE test run data reception deadline, GEBVs and file 734 should be sent to IDEA no later than 17:00 CET the Tuesday three (3) weeks after MACE test run data reception deadline. GMACE test run results will be distributed one (1) week after distribution of MACE test run results.
Truncated MACE: interested countries should submit their YYYY-4 data to the T-IDEA database no later than 17:00 CET the third Tuesday of October. Truncated Mace results will be distributed to participating countries three (3) weeks after.
Test results are confidential and are not meant to be published.
7.4 Test-run evaluation policy
7.4.1 Test-run evaluations intend to investigate the impact of new or modified national genetic evaluation results on the international evaluation; also to investigate potential improvements of the international genetic evaluation procedure. Results of such test-runs are reviewed by technical representatives of all countries involved. Pending on this review, changes are incorporated in the following routine evaluation.
7.4.2 Test-runs will be conducted in January, May (only for MACE) and September at the Interbull Centre. The January test-run intends to investigate changes with the view to introduce them in the April routine evaluation of the same year. The September test-run intends to investigate changes with the view to introduce them in the December routine evaluation of the same year. More in general, partecipation in a test evaluation run with modified data implies that an official implementation of the modification is targeted for a near future, usually within 6 months.
7.4.3 Participating countries are requested to consider potential changes/modifications of their national genetic evaluation procedure in line with the above-mentioned time schedule.
7.4.4 Guidelines for assessing the necessity of a test-run:
The table below presents the actions required/taken during a test and a routine run for Mace and Gmace:
MACE Test run |
1- Are any of the following rules applicable? (if not Go To 2) |
MACE Routine run |
5- Check the consistency of the data by the “verify” program |
GMACE Test run |
8- Check disparity between MACE and GMACE data |
GMACE Routine run |
10- |
1The rules for the traits to be validated are as before.
2 If the ΔSDg < 5%, the validation test is not needed.
7.4.4.1 Changes in models for genetic parameter and breeding value estimation and modification of pre-adjustment factors, such as age, require new validation according to the established procedure and a test-run before acceptance to a routine international evaluation.
7.4.4.2 Frequently countries introduce changes in their system with only minor effect on the national bull ranking. Such changes, however, may have considerable impact on international evaluations.One way to assess this would be to estimate the within-country sire variance with the procedure currently used by Interbull. Large changes of this parameter are invariably associated with substantial re-ranking in the international list.
7.4.4.3 Experience shows that changes in sire variance estimates (based on data included in the international evaluation) larger than 5% from one evaluation to the next can not be explained by added information from young bulls. In such case, validations and a test-run will be required.
8. Interbull CoP - Use of data for research
Upon specific agreement with participating organisations, the Interbull Centre can use data provided by these countries to conduct, on its own or in collaboration with other research institutions, research pertinent to international evaluations. Member organisations are encouraged to actively support such research and development projects, which will be under the auspices of the Interbull Steering Committee. A list of on going projects, own or collaborative, should be maintained by the Interbull Centre and made available on the Interbull home page at the Internet.
9. Interbull CoP - Promotion of services by the Interbull Centre
The Interbull Centre can promote the value of its services to potential users and supporters of the service with examples of results being produced. These results will be associated only with bulls officially proven in the respective countries of first sampling whose international predicted genetic merits have already been published.
10. Interbull CoP - Service fees
10.1 A running two-year budget for the service will be presented annually to the Interbull Steering Committee and member country representatives for approval.
10.2 Annual contributions to finance the service are based on a 5-Year Increase in Milk Daughters, and the current fee structure for dairy-production traits is as follows:
|
Production evaluation fee (€) |
|
Basic fee |
|
4 325.00 |
Variable fee, per 1 000 increased number of daughters (5-years period) |
≤ 100 |
53.05 |
101 to 300 |
20.23 |
|
301 to 1 000 |
7.57 |
|
1 001 to 2 400 |
5.09 |
|
> 2 400 |
0.28 |
|
Example: a country with a 5-years increase in number of daughters equal to 410 000 daughters will pay €4 325 (basic fee) + (100 × €53.05) + (200 × €20.23) + (109 × €7.57) = €14 501 for milk production traits.
All breeds are considered in the 5-Years Increase in Milk Daughters. The appropriate number of 5-years increase in milk daughters is computed by Interbull Centre based on the April's MACE production results of the year before.
For a country with more than one data provider for MACE services, the Basic Fee would be calculated using a 10% multiplier (4325 Euro x 10% = 433) that is added for each additional data-providing organization.
For any new country joining MACE services for production traits, the annual fee for the initial years shall be determined based on the No. of Milk Recorded Cows until the country has participated long enough for the fee to be calculated using the “5-Year Increase in Milk Daughters".
10.3 Interbull service fees for subscribing to the conformation traits evaluation are 30% of the service fee for milk production traits.
10.4 Interbull service fees for subscribing to the milk somatic cell and clinical mastitis evaluation are 15% of the service fee for milk production traits.
10.5 Interbull service fees for subscribing to the longevity evaluation are 15% of the service fee for milk production traits.
10.6 Interbull service fees for subscribing to the calving traits evaluation are 15% of the service fee for milk production traits.
10.7 Interbull service fees for subscribing to the female fertility traits evaluation are 20% of the service fee for milk production traits.
10.8 Interbull service fees for subscribing to the workability traits evaluation are 5% of the service fee for milk production traits.
10.9 Interbull service fees for subscribing to GEBV validation test are 15% of the service fee for milk production traits.
10.10 Interbull service fees for subscribing to multi-country genomic evaluations are described in Appendix VII
10.11 Interbull service fee for subscribing to the Truncated MACE evaluation are: a fixed fee of €1 500 for organisations submitting data for a single breed, and €2 500 for organisations submitting data for 2 or more breeds.The TMACE participation fee for a country with more than one data provider, is calculated using a 10% multiplier that is added for each additional data providing organisation
10.12 Sharing additional animal information via the IDEA AnimInfo module is currently free of charge.
10.13 Sharing of genetic traits information via IDEA AnimInfo is subjected to a fee as described in Appendix XI
10.14 Subscribing to the GenoEx-PSE service is subjected to a fee of €1030.
10.14 Fees apply to countries providing data for the international evaluation and also countries that are not prepared to provide data, but wish to receive international evaluation results.
10.15 Participating countries will be billed once per year, usually within 60 days after receiving the April international evaluation.
10.16 The official Interbull customer representative is responsible for raising the national contribution within country.
10.2 Service Fee Structure for novel traits
10.2.1 Interbull has proposed a funding model for novel traits that includes a fee structure expressed as a percentage of the fee paid for MACE services for production traits. This approach is consistent with all other trait groups with MACE services.
10.2.2 The distinction for novel traits, however, is that countries which have invested the longest on the collection of the new phenotype should benefit most from a lower fee. Countries with a higher proportion of bulls proven for the novel trait in question (i.e.: cma) relative to production (i.e.: Milk) can receive a level of credit to a maximum of 50%. Countries that have more recently started collecting the new phenotype should be given the opportunity to lower their fee after a reasonable period of collecting these phenotypes. Therefore, the period to be included in the fee calculation is set to a 15-year period, ending 6 years from the current year (i.e. 2015 in 2021).
The level of credit is determined by the proposed Contribution Category as shown in Table 1.
Table 1: cma Trait Data Contribution Categories
Contribution Category |
Min |
Max |
FEE |
1 |
0% |
19,9% |
100% |
2 |
20% |
39,9% |
87,50% |
3 |
40% |
59,9% |
75% |
4 |
60% |
79,9% |
62,50% |
5 |
80% |
100% |
50% |
10.2.3 Service Fee for cma MACE Services:
The above principles have led to the following cma MACE Service fee structure:
- The fees charged for cma MACE Services will be based on a 15-year window of birth years for proven bulls included in MACE for Milk. The last birth year to be included in the time window will be six years prior to the current calendar year (i.e.: ending in 2015 for 2021).
- The Contribution Categories applied to cma will be based on five groups by quintile, as outlined in Table 2.
- The “Base” fee for cma evaluations will be set at 8% of the Production fee. Therefore resulting in a fee of 4% for countries with the maximum credit based on Contribution Category 5 (80-100%).
- The fees for cma will be based solely on the calculations applied to the breed with the highest number of qualifying bulls for Milk, which will most often be the Holstein breed. Payment of such fees therefore allow that country to participate in the cma MACE services for all breeds.
- The Country of Origin for each bull will be designated as the country with the maximum number of daughters in the published MACE evaluation for Milk.
10.2.4 Fee calculations:
An example (estimate) of the fees for countries that are most likely able to join the new cma service is provided below.
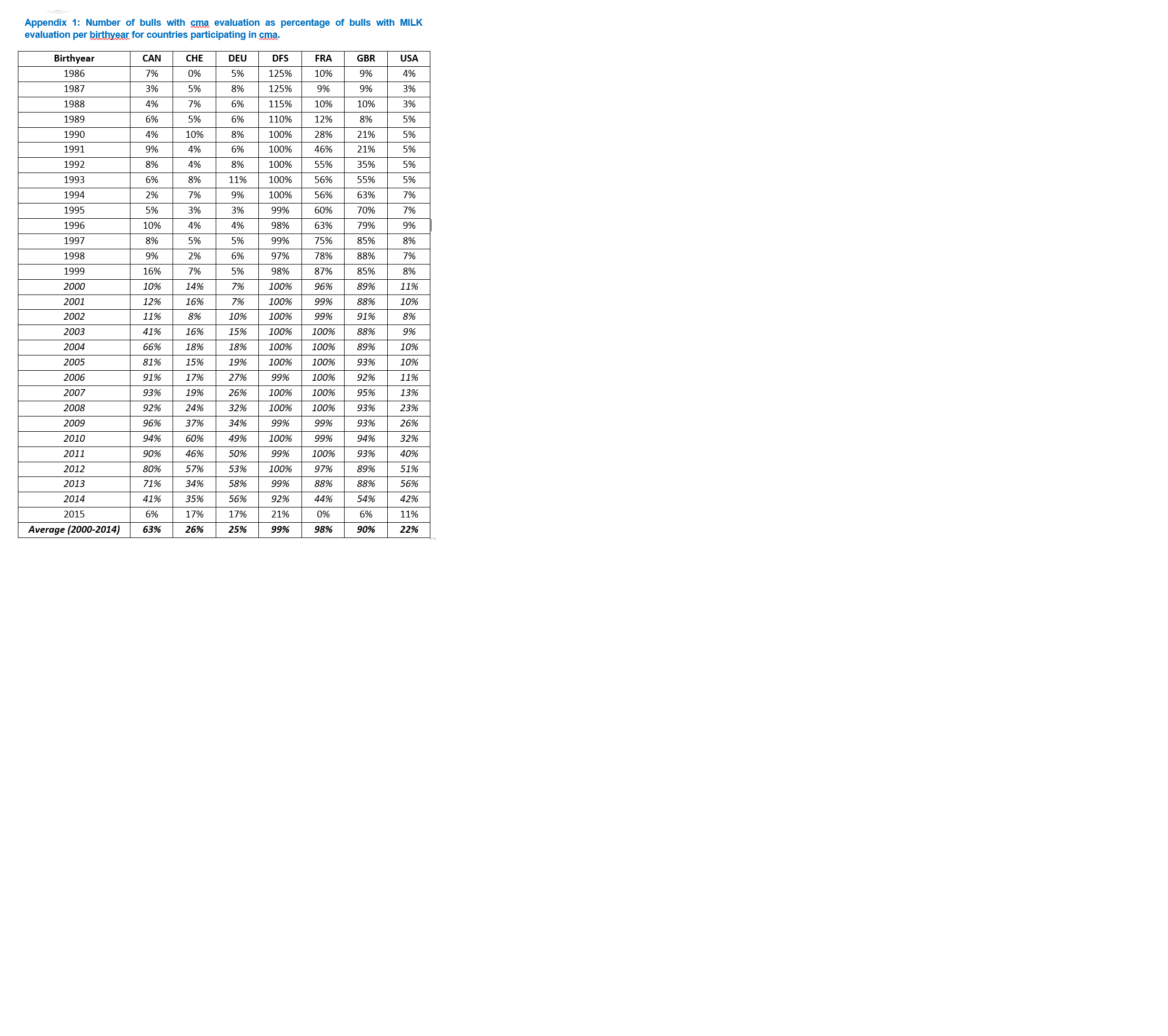
Proportion of bulls with a cma evaluation relative to MILK
- The proportion of bulls that have been evaluated for cma has been calculated on basis of the frequencies in individual years as defined above.
- Data was available for the period from 1986 until 2015 as shown in Appendix 1. The last row in Appendix 1 shows the average contribution in the 15-year period 2000-2014.
- This results in the cma data contribution categories, discounts and estimated fees as shown in Table 2.
Table 2: cma fee calculation by country
|
CAN |
CHE |
DEU |
DFS |
FRA |
GBR |
USA |
Average contribution 15-year period |
63% |
26% |
25% |
99% |
98% |
90% |
22% |
CMA data contribution category |
4 |
2 |
2 |
5 |
5 |
5 |
2 |
Discount |
37,5% |
12,5% |
12,5% |
50% |
50% |
50% |
12,5% |
CMA service fee (euro) |
771 |
956 |
1999 |
1010 |
966 |
692 |
1723 |
Interbull CoP - Appendix I - Service contracts and Letters of understanding
I-a: Service Contract - MACE evaluations
I-b: Letter of understanding - Conformation
I-c: Letter of understanding - Udder Health
I-d: Letter of understanding - Longevity
I-e: Letter of understanding - Calving
I-f: Letter of understanding - Female Fertility
I-g: Letter of understanding - Workability
I-h: Letter of understanding - SNP Training cma
I-i: Letter of understanding - AnimInfo
I-j: Service Contract - Genomic evaluations IG BSW
I-k: Service Contract - Genomic evaluation IG HOL
I-l: Letter of Agreement IG HOL
I-m:Letter of Agreement IG HOL contributing countries
I-n:Letter of understanding Genetic Traits Exchange
I-o: Letter of agreement for sharing of female imputed genotypes
Interbull CoP - Appendix II - Forms & Guidelines
Interbull CoP - APPENDIX III - Interbull trend validation procedures
Interbull Conventional Validation is carried out by a specific software, TrendTest software, available exclusively for the Interbull users and located in the IDEA database. The software carries out the three available methods according to the principle described below.
Mendelian Sampling Variance Test is carried out by a specific software available exclusively for the Interbull users and located in the IDEA database
Principle Before accepting data from a country to include in the international genetic evaluation, three trend validation tests are implemented. Data should pass all three tests, when applicable, in order to be included in the genetic evaluation of Interbull. Validation tests must be implemented when a country first enters the Interbull evaluation system and, thereafter, every time the country’s national genetic evaluation system is modified, or when the last validation was more than two years ago.
Description Development of the trend validation methods is credited to the scientific group of INRA in France. The following peer-reviewed publication provides deeper insight into the scientific principles on which the trend validation tests are based:
Boichard, D., Bonaiti, B., Barbat, A. and Mattalia, S. 1995. Three methods to validate the estimation of genetic trend for dairy cattle. J. Dairy Sci. 78, 431-437.
Weller, J. I., Emanuelson, U. and Ephraim, E. 2003. Validation of genetic evaluation methodology using the nonparametric Bootstrap Method. Interbull Bulletin 31, 26-29.
Lidauer, M., Mäntysaari, E.A., Pedersen, J. and Strandén. 2005. Model validation using individual daughter deviations – statistical power. Interbull Bulletin 33, 195-199.
A-M Tyrisevä, EA Mäntysaari, J Jakobsen, GP Aamand, J Dürr, WF Fikse, and MH Lidauer. 2018. Validation of consistency of Mendelian sampling variance. J Dairy Sci, 101:2187–2198.
A brief description of the three testing methods follows:
Testing Method 1
Definition: Comparison of genetic trends estimated using only first lactation versus all lactations in the routine national genetic evaluations.
Responsibility: Countries using multiple-lactation models in national genetic evaluations are required to apply this method prior to delivering data to the Interbull Centre.
Motivation: Investigate the impact of cow records from different age groups on the genetic trend.
Data: Data used in the routine national genetic evaluation, whose results will be included in the international evaluation. The same time edit as applied in the genetic evaluation of Interbull should therefore be used.
Action: Estimate the genetic trend in the AI bull population with the usual model (bT).
Compute the difference: = bT-bl, for each trait.
Criterion: | | must be less than .02*
g or .01*
g if breeding values or transmitting abilities were considered in the trend estimation, respectively;
g is the genetic standard deviation for the trait; when each lactation is treated as different trait, instead of
g consider (
g l*
gT)**.5/rglT, where
gl and
gT are the genetic standard deviations for first and all lactations (pooled) and rglT is the genetic correlation between first and all lactations (pooled).
Remarks: If b1<bT the genetic trend is over-estimated (the opposite if b1>bT). Possible solutions: reconsider age pre-adjustment factors, include an age by parity or age within parity effect in the model, and include parity in the definition of management (contemporary) groups.
Testing Method 2
Definition: Analysis of within bull yearly Daughter Deviations (e.g. Daughter Yield Deviations, DYD), hereafter referred to as DD.
Responsibility: Countries where DD are available are required to apply this method prior to delivering data to the Interbull Centre.
Motivation: DD are independent of the year of calving of bulls´ daughters. This method investigates the non-genetic time trend over the entire period considered in the national evaluation; deviations from zero will indicate biases in the genetic trend estimation.
Data: Within year DD computed in the most recent routine national genetic evaluation, whose results will be included in the international evaluations. All AI bulls first sampled in each country with daughters in at least 10 herds should be considered. For a test-day model individual daughter deviations are the sum of the additive genetic effect(s) of the sire, the Mendelian sampling term(s), the permanent environmental effect(s) pertaining to the daughter, and the residual term. A detailed description of computation of individual daughter deviations and (average) daughter deviations for test-day models is in Lidauer et al. (2005).
Action: Analyze DD with the following fixed effect model:
Yij=BULLi + b*j + eij [1]
where Yij is the DD considering daughters born in the jth year of the ith bull; by definition j=0 for the first year when at least 10 daughters of a bull were born; BULL is the effect of the ith bull.
Criterion: |b| from Model [1] must be less than .01* g, where
g is the genetic standard deviation for the trait.
Remarks: If b>0 the environmental trend is under-estimated and the genetic trend is over-estimated (the opposite if b<0)
Testing Method 3
Definition: Analysis of official national predicted genetic merit variation across evaluation runs.
Responsibility: Countries that can not implement Method 1 and 2 due to model restrictions are required to apply this method prior to delivering data to the Interbull Centre. Countries that have applied both Methods 1 and 2, may also apply this method if they wish, but it is not required. The Interbull Centre implements this method with data from ALL participating organisations. Each participating organisation is responsible for delivering appropriate data to the Interbull Centre.
Motivation: Investigate the random variation associated with new daughters. The method is independent of country and can be applied at the venue of the international evaluations.
Data: Let YYYY be the year of the most recent national genetic evaluation. Data include bulls born between certain years (see section Time windows below), AI first sampled in each country, whose national genetic evaluation of year YYYY-4 was based only on first crop daughters (minimum 20 daughters in 10 herds). For each one of these bulls the following variables are needed:
X = National evaluation (predicted genetic merit) from the new model using the same data as in the last routine genetic evaluation run in year YYYY-4;
Y = National evaluation (predicted genetic merit) from the new model using the data from the most recent routine genetic evaluation run, whose results will be included in the international evaluation (year YYYY);
nx= Total number of daughters on which X was based in year YYYY-4;
nY= Total number of daughters on which Y was based in year YYYY;
nj = Number of new (first calving) daughters considered in the last national evaluation of the jth year, where j=1 for YYYY-3, j=2 for YYYY-2, j=3 for YYYY-1 and j=4 for YYYY;
t = Proportion of bull´s new daughters, where
where mj= (YYYY-4) + j - (mean year of first calving of daughters on which the bull´s national evaluation in year YYYY-4 was based).
Time window: Appropriate time windows (birth years of bulls) may vary depending of the trait to be validated, the speed of their progeny test program and other factors. A shift of the time window with one year will give a different set of bulls that qualify for the test. It is desirable to choose a time window so as to have the largest number of qualifying bulls as possible. It is required that bulls do have only first crop progeny for the evaluation in (YYYY - 4), and first and second crop progeny for the evaluation in YYYY. To fulfill these requirements for calving traits birth years commonly chosen are between YYYY-10 and YYYY-7 for direct calving ease / stillbirth), between YYYY-12 and YYYY-9 for maternal calving ease / maternal stillbirth and between YYYY-14 and YYYY-10 for production traits.
Action: Analyse Y for the ith bull with the following model: Yi = a + b*Xi + *ti + ei [2] weighing each observation by w w=Weight of Y, where
where: k=variance ratio or k= , h2=heritability of the trait
R=expected correlation between methods used for calculation of Y and X (R = .99); h2 should be provided by each country.
Precision: The empirical 95% confidence interval of should be submitted with the validation results. The empirical confidence interval can be obtained by a non-parametric bootstrap (Weller et al, 2003): Repeat at least 1000 times:
i. Sample (with replacement) among qualifying bulls. Sample size is the same as the number of qualifying bulls;
ii. Compute the regression coefficient ( ) for this sample. The 95% empirical confidence interval is obtained as the 2.5% and 97.5% percentile values of computed under ii.
Criterion: If from Model [2] differs significantly from zero (zero is not within the empirical 95% confidence interval) then |
| from Model [2] must be less than .02*
g or .01*
g if breeding values or transmitting abilities were considered in the trend estimation, respectively, where
g is the genetic standard deviation for the trait.
Remarks: If >0 the genetic trend is over-estimated (the opposite if <0).
LogisticsThe three testing methods must be implemented before a country sends data for an Interbull test-run. Results must be made available to the Interbull Centre on specifically designed forms that each country receives upon subscribing to the Interbull service. Data for testing method 3 are sent from each country to the Interbull Centre according to a particular format that is described in the service package that each country also receives upon subscribing to the Interbull service.
When data from a country fail a test Possible reasons for failing a validation test are
1) problems with the national genetic evaluation model in a country;
2) sampling error;
3) test is not appropriate for the genetic model in question.
The Interbull Centre investigates results in order to establish their statistical significance. With small data sets, discrepancies may sometimes not be statistically significant. In such cases, reasons for failing the validation tests are attributable to chance. When results fail a validation test, and deviations from the established criteria are statistically significant, it is assumed that something is not completely correct with the national genetic evaluation model. Interbull Centre is then contacting this country for an effort to resolve the problem. In such cases, the aim is to revise the national genetic evaluation model so that results comply with the validation tests.
Testing Method IV - Mendelian Sampling Variance Test
Definition: Estimates within-year genetic variances and tests for a possible trend and outliers of the estimated variances
Responsibility: MS test results are not used by Interbull Centre as a requirement for participating in MACE evaluation services, but are considered as a great investigative tool for National Genetic Centers to detect possible areas requiring improvement in their models or data structures.
Motivation: Test the homogeneity of variance across years since under- or overestimation of genetic variance in some country affects the spread of breeding values on the other country scales, which can significantly affect the ranking of top bulls.
Data: Animal’s birth year, EBVs for the animals and their parents and the estimates of the EBV reliabilities. Only animals with complete information on both parents should be included in the analyses. MS test should be applied to both males and females but application of the test to females should be limited to medium-high heritability traits (usually production, stature and somatic cell score). Lower heritability traits should be tested on males only. MS test should be applied to the same traits for which conventional validation is conducted. This implies that for conformation traits the test should be applied to the following traits only: STA, USU, LOC and BCS. For bulls, the same data edits should be applied as outlined in the Interbull Code of Practice, item 5.1.3. No specific data edits are required for females.
Actions A complete description of the validation procedure is available here
Interbull CoP - APPENDIX IV - Description of weighting factors and examples
Weighting factors for the international genetic evaluation
The following is a procedure to compute new weighting factors for the international genetic evaluation of Interbull, and has to be implemented separately by each individual organization participating in Interbull evaluations. The procedure is based on information used in the national genetic evaluation in each country, and how a country considers such information depends on the genetic evaluation model.
The procedure consists of two steps. In the first step, for each animal with own performance records used in the national genetic evaluation, the reliability due to its own performance, R(o), is estimated using selection index methodology. R(o) is computed separately per trait (milk, fat or protein yield) and information from the other traits in a multiple trait national genetic evaluation is ignored. The second step combines the R(o) from the daughter and her dam, expressed in effective daughter contributions (EDC), which are subsequently accumulated over all daughters of a sire.
Step 1: Estimation of reliability based on own performance (R(o)):
For each animal with own performance records in the national genetic evaluation, Ri(o) is estimated. Estimation of Ri(o) depends on the genetic evaluation model.
a) Single trait (repeatability) model for the national genetic evaluation (where individual lactations are considered as the same trait):
b) Multiple trait model for the genetic evaluation (where each lactation, part of lactation or test day observation are treated as different traits):
Let k’EBV be the estimated breeding value or transmitting ability of the bull for the trait of interest (milk, fat or protein yield), i.e. the one submitted in the 010 file to Interbull, where EBV is a vector with multiple trait (lactation, part-lactation, test day) estimates of breeding value or transmitting ability, and k a vector with weights given to each estimate.
Note:
- heritability is the heritability of a single observation, e.g. test day yield - non-genetic parameters correspond to all non-genetic effects, which may include permanent environment effects ( i.e. E = PE + e )
- the non-genetic correlation depends on assumptions of the evaluation model, i.e. whether PE and/or e are considered to be correlated between lactations
- in case genetic and environmental correlations and heritability are not constant over lactation (e.g. in random regression models), an average value over lactation should be used
for missing traits, computation of the nominator in (2) can be done in two ways:1) set the rows and columns corresponding to the missing trait to zero in the P matrix, or 2) remove the corresponding rows and columns in the P matrix and the corresponding rows in the G matrix. Either way should give the same results. However, the first method is recommended since it is less ambiguous (i.e. the G matrix is not affected and the P matrix is of same dimension under all circumstances). The denominator in (2) is the same for all animals, whether observations are missing or not!
Step 2: Combine sources of information
a) Once Ri(o) is computed for all animals with own records in the genetic evaluation, information from the cow and her dam is combined as follows:
This new weight, resulting from Equation (4), will be added to the trait-group files according to the revised format and sent to Interbull.
References
Fikse, W.F. and Banos, G., 1999. Weighting factors of daughter information in international genetic evaluation for milk production traits: effect on (co)variance components. J. Dairy Sci., 82 (suppl. 1): 72 (Abstr.).
Fikse, W.F. and Banos, G., 1999. Weighting factors in international genetic evaluations: effects on international breeding value and reliability estimates. Interbull Bulletin, 22: 38 - 43.
Fikse, W.F. and Banos, G., 2001. Weighting factors of sire daughter information in international genetic evaluations. J. Dairy Sci. 84:1759-1767.
VanRaden, P.M. and Wiggans, G.R., 1991. Derivation, calculation, and use of national animal model information. J. Dairy Sci., 74: 2737 - 2746.
VanVleck, L.D., 1993. Selection index and introduction to mixed model methods. pp 481. CRC Press Inc., Boca Raton, FL.
Weighting factors for the international genetic evaluation of longevity
The following describes the procedure to compute weighting factors for the international genetic evaluations for longevity traits, and has to be implemented by each country participating in Interbull evaluations for longevity traits. Procedures currently in use for genetic evaluation of longevity traits can be divided into two main categories: survival analysis and analysis of culling rate, stayability or length of life with mixed linear models. Due to the different nature of both types of analysis, procedures to compute weighting factors are described separately.
Survival analysis²
The number of culled daughters is to be submitted as weighting factor. Heritability should be computed as:
Mixed linear model analysis
- If longevity data are analysed with mixed linear models, assuming normality of breeding values and residuals, then the procedures above underneath “New weighting factors for the international genetic evaluation" applies.
References Ducrocq, V., Delaunay, I., Boichard, D. and S. Mattalia. 2003. A general approach for international genetic evaluations robust to inconsistencies of genetic trends in national evaluations. Interbull Bulletin 30, 101-111.
²A more elaborate definition of a weighting factor for survival analysis can be found in Ducrocq et al. (2003). The weighting factor proposed there was observed to have a perfect correlation with number of culled daughters (Ducrocq, 2003, pers. comm.), why the latter definition of weighting factors will be used for international evaluations
Weighting factors, example
Weighting factors for the international genetic evaluation: Example
This document is a supplement to Appendix IV (which will be referred to as the “parent” document) and contains a set of examples to illustrate the calculation of the new weighting factors. For a hypothetical data set, Step 1 of the procedure will be illustrated for two different types of national genetic evaluation models. Computations for Step 2 are then outlined for a given set of results from Step 1.
Step 1: Computation of reliability based on own performance (R(o)): Consider the following hypothetical data set:
Animal |
Sire |
Lactation 1 |
Lactation 2 |
||
|
|
CG |
Weight1 |
CG |
Weight1 |
1 |
S1 |
A1 |
1 |
B1 |
1 |
2 |
S1 |
A1 |
1 |
B1 |
1 |
3 |
S1 |
A1 |
1 |
B1 |
1 |
4 |
S2 |
A1 |
1 |
B1 |
1 |
5 |
S2 |
A1 |
1 |
B1 |
0.9 |
6 |
S2 |
A1 |
1 |
B2 |
1 |
7 |
S1 |
A1 |
0.85 |
- |
- |
8 |
S1 |
A1 |
0.7 |
- |
- |
9 |
S1 |
A2 |
1 |
B2 |
1 |
10 |
S2 |
A2 |
1 |
B2 |
1 |
11 |
S2 |
A2 |
1 |
B2 |
1 |
12 |
S2 |
A2 |
1 |
B2 |
1 |
13 |
S1 |
A2 |
1 |
B2 |
1 |
14 |
S1 |
A2 |
1 |
B2 |
0.75 |
15 |
S1 |
A2 |
0.8 |
- |
- |
1 Weight in the national genetic evaluation. Computation of Ri(o) depends on the genetic evaluation model, and will for this example data set be illustrated for two classes of models.
a) Single trait (repeatability) model for the national genetic evaluation (where individual lactations are considered as the same trait):
Assume the following parameters from the national genetic evaluation: h2 = 0.30 r = 0.50
For animals 1-8 in lactation 1:
Likewise, for animals 9-15 in lactation 1, animals 1-5 in lactation 2, and animals 6 and 9-14 in lactation 2 the summation evaluates to 6.8, 4.9, and 6.75, respectively.
For animals 1-3 and 7-8 in lactation 1:
This summation evaluates to 3.0, 3.8, 3.0, 3.0, 1.9, 4.0, and 2.75 for animals 4-6 in lactation 1, animals 9 and 13-15 in lactation 1, animals 10-12 in lactation 1, animals 1-3 in lactation 2, animals 4-5 in lactation 2, animals 6 and 10-12 in lactation 2, animals 9 and 13-14, respectively. Then for animal 1:
For animals 1 - 15 the results are:
Animal |
wi1 |
wi2 |
m |
Ri(o) |
1 |
0.397 |
0.388 |
0.785 |
0.264 |
2 |
0.397 |
0.388 |
0.785 |
0.264 |
3 |
0.397 |
0.388 |
0.785 |
0.264 |
4 |
0.603 |
0.612 |
1.215 |
0.329 |
5 |
0.603 |
0.551 |
1.154 |
0.321 |
6 |
0.603 |
0.407 |
1.010 |
0.302 |
7 |
0.338 |
- |
0.338 |
0.152 |
8 |
0.278 |
- |
0.278 |
0.131 |
9 |
0.441 |
0.593 |
1.034 |
0.305 |
10 |
0.559 |
0.407 |
0.966 |
0.295 |
11 |
0.559 |
0.407 |
0.966 |
0.295 |
12 |
0.559 |
0.407 |
0.966 |
0.295 |
13 |
0.441 |
0.593 |
1.034 |
0.305 |
14 |
0.441 |
0.444 |
0.886 |
0.282 |
15 |
0.353 |
- |
0.353 |
0.157 |
b) Multiple trait model for the national genetic evaluation (where each lactation, part lactation or test day observation is treated as a different trait):
The same data structure as before is used in this case too, however, the two lactations are considered as genetically distinct traits. The example calculations below assume only one observation per trait. mj for animal i is therefore equal to wij as computed above. The complete matrix of phenotypic (co)variances that is used in the national genetic evaluation will be denoted as P* for the remaining of this document. The matrix specific to each animal (with appropriate elements set to zero) will be denoted as P. The illustration of missing observations (animal 7) is shown for the first implementation option, i.e. zeroing rows and columns, since this method is less ambiguous (i.e., the G matrix is not affected and the P matrix is of same dimension under all circumstances).
Assume the following parameters from the national genetic evaluation:
For animals 1 - 15 the results are:
Animal |
m1 |
m2 |
nominator |
Ri(o) |
1 |
0.397 |
0.388 |
21.673 |
0.170 |
2 |
0.397 |
0.388 |
21.673 |
0.170 |
3 |
0.397 |
0.388 |
21.673 |
0.170 |
4 |
0.603 |
0.612 |
30.954 |
0.242 |
5 |
0.603 |
0.551 |
29.423 |
0.230 |
6 |
0.603 |
0.407 |
25.952 |
0.203 |
7 |
0.338 |
- |
9.487 |
0.074 |
8 |
0.278 |
- |
7.813 |
0.061 |
9 |
0.441 |
0.593 |
28.176 |
0.221 |
10 |
0.559 |
0.407 |
25.143 |
0.197 |
11 |
0.559 |
0.407 |
25.143 |
0.197 |
12 |
0.559 |
0.407 |
25.143 |
0.197 |
13 |
0.441 |
0.593 |
28.176 |
0.221 |
14 |
0.441 |
0.444 |
24.025 |
0.188 |
15 |
0.353 |
- |
9.914 |
0.078 |
In case multiple observations are recorded on genetically distinct traits, the P11 element in the P matrix is obtained as follows:Let two observations be recorded on the first genetically distinct trait, the repeatability of observations on the same trait r1 = 0.40. Assume that wi1, computed the same way as illustrated above, for observation 1 and 2 are 0.7 and 0.8. Then m1 = 0.7 + 0.8 = 1.5, and
Step 2: Combine sources of information
First, the reliability contributed by the dam of the animal is added to the animals reliability based on own performance, and expressed in effective daughter contributions (EDC) (formula (3) in the “parent” document). Once that is done for all animals, information from all daughters of a sire is accumulated into one single weighting factor for each sire. Ri(o) values from case b) from Step 1 will be taken for these example computations, and hypothetical values for Rdam(o) are used. Parameters (G, P*, k) will also be those used in case b) of Step 1.
Note that Rdam(o) is always zero in case the national genetic evaluation applies a sire model!
The heritability of the trait submitted in the 010 file is the heritability needed in Step 2, and is for this example computed as:
For animals 1 - 15 the EDCi(o+d), given the assumed values for Rdam(o) for dams, are:
Animal |
Sire |
Ri(o) |
Rdam(o) |
EDCi(o+d) |
1 |
S1 |
0.170 |
0.29 |
0.486 |
2 |
S1 |
0.170 |
0.30 |
0.486 |
3 |
S1 |
0.170 |
- |
0.479 |
4 |
S2 |
0.242 |
0.31 |
0.712 |
5 |
S2 |
0.230 |
0.31 |
0.674 |
6 |
S2 |
0.203 |
0.29 |
0.588 |
7 |
S1 |
0.074 |
0.28 |
0.206 |
8 |
S1 |
0.061 |
0.26 |
0.169 |
9 |
S1 |
0.221 |
0.24 |
0.640 |
10 |
S2 |
0.197 |
0.23 |
0.567 |
11 |
S2 |
0.197 |
0.23 |
0.567 |
12 |
S2 |
0.197 |
0.23 |
0.567 |
13 |
S1 |
0.221 |
0.24 |
0.640 |
14 |
S1 |
0.188 |
0.25 |
0.541 |
15 |
S1 |
0.078 |
0.27 |
0.215 |
Finally, the weighting factors ws for both sires in this example data set are computed as follows:
Weighting factors for the international genetic evaluation of longevity:
Examples for mixed linear models
This document contains a set of examples to illustrate the calculation of the weighting factors for genetic evaluations for longevity using mixed linear models. The same procedure will apply as the one for production traits (outlined in Appendix IV). It consists of two steps:
1) computation of reliability based on own performance, and
2) combining information from daughters and their dams, to be expressed in effective daughter contributions (EDC). For a hypothetical data set, computations for Step 1 will be illustrated for two different national genetic evaluation models based on mixed linear models. Computations for Step 2 are then outlined for a given set of results from Step 1.
Step 1: Computation of reliability based on own performance (R(o)):
Consider the following hypothetical data set:
Animal |
Sire |
CG |
Length of life analysis |
|
Binomial trait analysis |
|
|
|
|
Weight1 |
Productive life (months) |
|
Survived 2nd lactation (0=no, 1=yes) |
1 |
S1 |
A1 |
1 |
23 |
|
0 |
2 |
S1 |
A1 |
1 |
29 |
|
1 |
3 |
S1 |
A1 |
1 |
24 |
|
1 |
4 |
S2 |
A1 |
1 |
23 |
|
0 |
5 |
S2 |
A1 |
1 |
28 |
|
1 |
6 |
S2 |
A1 |
1 |
24 |
|
1 |
7 |
S1 |
A1 |
0.67 |
20 |
|
0 |
8 |
S1 |
A1 |
0.80 |
26 |
|
1 |
9 |
S1 |
A2 |
1 |
28 |
|
1 |
10 |
S2 |
A2 |
1 |
18 |
|
0 |
11 |
S2 |
A2 |
1 |
26 |
|
1 |
12 |
S2 |
A2 |
1 |
23 |
|
0 |
13 |
S1 |
A2 |
1 |
28 |
|
1 |
14 |
S1 |
A2 |
1 |
24 |
|
1 |
15 |
S1 |
A2 |
0.81 |
27 |
|
1 |
1 Weight in the national genetic evaluation. Computation of Ri(o) depends on the genetic evaluation model, and will for this example data set be illustrated for two types of linear model analysis. For the first case, longevity is regarded as a binomial trait indicating whether or not an animal survived the first and second lactation. For the second case, length of productive life is recorded. In the example a situation is imitated where some records receive a lower weight (e.g., due to those being predicted rather than observed records).
a) Binomial trait analysisAssume the following parameters from the national genetic evaluation: h2 = 0.02
For animals 1-8:
Likewise, for animals 9-15 the summation evaluates to 7.
Then for animal 1:
For animals 1 - 15 the results are:
Animal |
m = wi1 |
Ri(o) |
1 |
0.375 |
0.0075 |
2 |
0.375 |
0.0075 |
3 |
0.375 |
0.0075 |
4 |
0.625 |
0.0125 |
5 |
0.625 |
0.0125 |
6 |
0.625 |
0.0125 |
7 |
0.375 |
0.0075 |
8 |
0.375 |
0.0075 |
9 |
0.429 |
0.0086 |
10 |
0.571 |
0.0114 |
11 |
0.571 |
0.0114 |
12 |
0.571 |
0.0114 |
13 |
0.429 |
0.0086 |
14 |
0.429 |
0.0086 |
15 |
0.429 |
0.0086 |
b) Length of life analysis
The same data structure as before is used here as well, however, the trait being analysed is length of productive life. In addition, a situation with differential weights for observations is illustrated. Assume the following parameters from the national genetic evaluation: h2 = 0.10
For animals 1-8:
Likewise, for animals 9-15 the summation evaluates to 6.81.
Then for animal 1:
For animal 7:
- For animals 1 - 15 the results are:
Animal |
m1 |
Ri(o) |
1 |
0.402 |
0.040 |
2 |
0.402 |
0.040 |
3 |
0.402 |
0.040 |
4 |
0.598 |
0.060 |
5 |
0.598 |
0.060 |
6 |
0.598 |
0.060 |
7 |
0.269 |
0.027 |
8 |
0.321 |
0.032 |
9 |
0.441 |
0.044 |
10 |
0.560 |
0.056 |
11 |
0.560 |
0.056 |
12 |
0.560 |
0.056 |
13 |
0.441 |
0.044 |
14 |
0.441 |
0.044 |
15 |
0.357 |
0.036 |
Step 2: Combine sources of information
First, the reliability contributed by the dam of the animal is added to the animals reliability based on own performance, and expressed in effective daughter contributions (EDC). Once that is done for all animals, information from all daughters of a sire is accumulated into one single weighting factor for each sire. Ri(o) values from case b) from Step 1 will be taken for these example computations, and hypothetical values for Rdam(o) are used.
Note that Rdam(o) is always zero in case the national genetic evaluation applies a sire model!
Assume the following parameters from the national genetic evaluation: h2 = 0.10 and k follows as:
For animal 1:
And for animal 3, which is assumed to have an unknown dam
R3(o) = 0.040
Rdam(o) = 0
For animals 1 - 15 the EDCi(o+d), given the assumed values for Rdam(o) for dams, are:
Animal |
Sire |
Ri(o) |
Rdam(o) |
EDCi(o+d) |
1 |
S1 |
0.040 |
0.059 |
0.394 |
2 |
S1 |
0.040 |
0.060 |
0.394 |
3 |
S1 |
0.040 |
- |
0.394 |
4 |
S2 |
0.060 |
0.062 |
0.594 |
5 |
S2 |
0.060 |
0.062 |
0.594 |
6 |
S2 |
0.060 |
0.058 |
0.594 |
7 |
S1 |
0.027 |
0.056 |
0.265 |
8 |
S1 |
0.032 |
0.052 |
0.315 |
9 |
S1 |
0.044 |
0.048 |
0.434 |
10 |
S2 |
0.056 |
0.046 |
0.554 |
11 |
S2 |
0.056 |
0.046 |
0.554 |
12 |
S2 |
0.056 |
0.046 |
0.554 |
13 |
S1 |
0.044 |
0.048 |
0.434 |
14 |
S1 |
0.044 |
0.050 |
0.434 |
15 |
S1 |
0.036 |
0.054 |
0.354 |
Finally, the weighting factors ws for both sires in this example data set are computed as follows:
Interbull CoP - APPENDIX VI - Interbull Evaluation Pre-Release Agreement
Between The Interbull Centre
And
as one of the Interbull member organisations receiving international bull evaluation results, (hereinafter referred to as “The Member”) ,
WHEREAS The Interbull Centre recognizes the potential benefits associated with an extended review of the international bull evaluations prior to their official release to all participating countries; and
WHEREAS The Member has interested personnel with the expertise required to analyze international bull evaluations as computed by The Interbull Centre; and
WHEREAS The Member also recognizes the benefits associated with the receipt, analysis and processing of international bull evaluation results prior to their official release;
THEREFORE, the parties agree to the following:
1.0 That The Interbull Centre will provide to The Member all required electronic files and pertinent documents related to the international bull evaluations for all traits and breeds in accordance with the target time and date of 12:00 CET on the Thursday prior to each Tuesday associated with a scheduled official release.
2.0 That The Member accepts and respects the desired level of confidentiality associated with the early receipt of the international bull evaluation results, either implicitly or explicitly, recognizing that the sole intent of the pre-release by The Interbull Centre is the validation of results and the possibility of pre-processing by The Member prior to official release, no earlier than 07:00 local time on the Tuesday of targeted official release. It is the responsibility of the signing authority representing The Member in this agreement to ensure that the use of the international bull evaluations respects the intent of this agreement.
3.0 That The Interbull Centre will provide a status report, usually via an e-mail message, to the signing authority, or their assigned delegate representing The Member, prior to a target time and date of 17:00 CET on the Friday following the pre-release of the international bull evaluations, including an indication of the likelihood that the Tuesday morning official release will be respected for official release by the participating countries.
4.0 That The Interbull Centre will provide this service, at no additional charge, to The Member that is in good standing with regards to the payment of regular Interbull fees and The Interbull Centre is authorized to cease provision of the service to any organisation, if regular payment of Interbull fees is not carried out.
5.0 Breach of clause 2.0 of this agreement by The Member, as determined by The Interbull Centre in a fair and reasonable manner, will result in the following actions regarding the possible termination of the service to The Member:
- 5.1 The Interbull Centre will inform The Member in writing of the specific breach requesting that it be rectified within a period of 30 days to the satisfaction of The Interbull Centre. Specific actions taken by The Member to avoid future breaches of this agreement must be communicated in writing to The Interbull Centre.
- 5.2 In the event that The Member does not rectify a breach as per clause 5.1, or in the event of multiple breaches by The Member, The Interbull Centre will bring a recommendation to the Interbull Steering Committee for the termination of this agreement for a period of time to be determined by the Interbull Steering Committee.
6.0 This agreement takes effect with the international bull evaluation release scheduled for Monday August 12, 2002 or following the date of signing below, whichever is latest. It is not limited in time and can be modified or terminated upon mutual written consent from both parties. The Interbull Steering Committee reserves the right to terminate this service and/or agreement as needed.
Signed by: On behalf of The Member;
(Name of organisation receiving international bull evaluation results)
(Name of authorized representative)
(Signature of authorized representative and Date)
AND
On behalf of the Interbull Centre;
(Interbull Centre Director and Date)
Interbull CoP - APPENDIX VII- Annual service fees for genomic evaluations
Table 1 – Service fees for the international genomic evaluation: strata and respective fee per 1000 cows in milk recording scheme.
Minimum |
Maximum |
Fee per 1000 milk recorded cows |
Cumulative per stratum |
1 |
50000 |
51.00 € |
2550 € |
50001 |
100000 |
20.40 € |
3570 € |
100001 |
200000 |
10.20 € |
4590 € |
200001 |
|
5.10 € |
|
Table 2 – Examples of annual fees (EUR) per country for the international genomic evaluation composed by a fixed base (€2500) plus a variable part related to the size of the cow population.
Country |
Number of cows [1] |
Base |
Variable part [2] |
Base+Variable |
Austria-Germany |
226377 |
2 500 € |
4 632 € |
7 132 € |
France |
17430 |
2 500 € |
872 € |
3 372 € |
Italy |
98204 |
2 500 € |
3 464 € |
5 964 € |
Slovenia |
11200 |
2 500 € |
560 € |
3 060 € |
Switzerland |
175629 |
2 500 € |
4 256 € |
6 756 € |
USA |
17765 |
2 500 € |
888 € |
3 388 € |
Total |
546605 |
15 000 € |
14 672 € |
29 672 € |
- [1] Number of Brown Swiss milk recorded cows provided by the World Brown Swiss Federation.
- [2] Based on Table 1.
Interbull CoP - Appendix VIII - Interbull validation test for genomic evaluations - GEBV test
Document based on:
Sullivan, P.G. 2023. Updated Interbull software for genomic validation tests. Interbull Bulletin 58, p.7-16.
VanRaden, P.M. 2021. Improved genomic validation including extra regressions. Interbull bulletin 56: 65-69.
Mäntysaari, E., Liu, Z and VanRaden P. 2010. Interbull Validation Test for Genomic Evaluations. Interbull Bulletin 41, p. 17-21.
Definitions:
- EBV - Estimated Breeding Value (conventional national evaluations of the trait, free of genomic information, which are submitted to Interbull to be used in MACE evaluations)
- DGV - Direct Estimated Genomic Value (genomic evaluations based on SNP prediction equations)
- GEBV - Genomically Enhanced Estimated Breeding Value (evaluations that combine EBV and DGV)
- dGEBV - De-regressed GEBV
- GREL - Genomic reliability of the bull's GEBV
- EDC - Effective Daughter Contribution
- MACE - Multiple Trait Across Country Evaluation
- PA - Parent Average
- DD - Daughter Deviation
- NGEC - National Genetic Evaluation Centre
Motivation
The inclusion of national genomic information in international comparisons for dairy breeds requires that the national genomic breeding values (GEBV) get validated by Interbull in a similar fashion that conventional EBV are validated as a pre-condition to participate in the MACE evaluations.
The GEBV test will be applied to validate national models used to compute GEBV that the national genetic evaluation centers (NGEC) publish and will eventually submit to Interbull for international genetic evaluations including genomic information. The GEBV test can also be considered a quality assurance assessment for national genomic evaluations. GEBV from models that have been tested can be referred to as breeding value estimates with appropriate reliability, and which can be converted to other country scale breeding values using conversion equations derived by Interbull.
Rationale
The GEBV test evaluates:
- the unbiasedness of the genomic evaluations through the evaluation of
- the consistency of the genetic trend captured by GEBV,
- the consistency of bull rankings before versus after having progeny, and
- the consistency of the variation of GEBV relative to EBV;
- the improvement in selection accuracy from the use of GEBV instead of EBV.
A time-oriented cross-validation is used to test how well genomic evaluations of young bull calves, using current models and phenotypic data from 4 years ago, can predict current progeny performance. The NGEC shall re-run their current evaluation software while excluding the most recent 4 years of daughter phenotypes, to obtain reduced-data genetic (EBVr) and genomic (GEBVr) evaluations. The software will then test if the ranking and variance of bull GEBVr match statistical and genetic expectations relative to ranking and variance of the bull comparisons based on current progeny differences, as an indication of unbiasedness. Furthermore, if the GEBVr are more highly correlated than EBVr with the current progeny phenotypes, it is an indication of accuracy improvement with GEBV.
Linear regression models are used for the validation test, where the expected value of regression slopes equals 1 if validation bulls are an unselected group, and a value less than 1 if only a selected subgroup of the most recent proven bulls have been genotyped. The expected slope is lower with selective genotyping due to effects of selection on variances and covariances used to compute the validation slope. The software will account for effects of selective genotyping on expected slopes, using estimates of selection differential from the differences between average EBV of the genotyped bulls versus all proven bulls born in the period considered for validation testing. Bootstrapping is used for all significance testing, and a combination of statistical and biological limits of tolerance is used by Interbull to assign an overall assessment of pass or fail.
Test data sets
Data formats are described at https://interbull.org/ib/gebvtest_software_2024
Full data sets
Two sets of currently official evaluations for progeny-proven bulls shall be provided for the GEBV test. These will be the EBV and GEBV published or otherwise indirectly used by the NGEC for national selection programs. All bulls provided to Interbull in file300 for MACE shall be included in a conventional EBV file (file300Cf) for the GEBV test, and all these same bulls who are genotyped and have a national GEBV shall be included in the GEBV file (file300Gf).
Conventional national genetic evaluation file (file300Cf)
The national EBV sent by the NGEC as input for the most recent Interbull MACE evaluation will be used to identify validation test candidate bulls, estimate the intensity of selective genotyping, and check bulls birth year and type of proof.
Official national genomic evaluation file (file300Gf)
The national GEBV of current MACE bulls will be used to derive target values reflecting unbiased estimates of average progeny performance for the validation test bulls. The official validation target is derived internally by the software, based on the consistent application for all NGEC of a standardized international method for dGEBV developed by VanRaden(2021).
Reduced data sets
The reduced data sets should be prepared by truncating the phenotypes used as input for both the conventional and the genomic evaluations. The NGEC must exclude phenotypic information from the most recent 4 years and re-run the current models of genetic and genomic evaluation for the traits of interest. The pedigree should not be truncated, just the phenotypes, because each validation bull's predicted genetic contributions in future progeny, based solely on the bull's parent average (EBVr=PA) and on PA plus genomic prediction equations (GEBVr) from the reduced-data evaluations will be needed for the validation test.
Reduced conventional genetic evaluation file (file300Cr)
The NGEC shall carry out a conventional genetic evaluation with no genotypes, while using the truncated data (only phenotypes up to 4 years prior to the date of analysis) but including in the analysis all animals present in the current official evaluations used in MACE (file300Cf).
A minimum of 10 most recent birth years of proven bulls included in file300Cf must also be included in file300Cr. The older proven bulls, with progeny proofs already in the reduced data, are required as a comparative control group, to contrast evaluation changes for younger bulls in the validation test group relative to the older control bulls.
Reduced genomic evaluation file (file300Gr)
The NGEC shall carry out a genomic evaluation that includes the genotypes, while using the truncated data (only phenotypes up to 4 years prior to the date of analysis) but including in the analysis all animals present in the current official evaluations used as input to MACE (file300Cf). All bulls included in the conventional file300Cr who are also genomically evaluated must be included in the genomic file300Gr.
If a significant number of foreign bulls are included in the reference population for national genomic evaluations, and estimations of genomic prediction equations use de-regressed MACE values for these bulls as input, the reduced genomic evaluation can be achieved in three ways, listed by descending order of preference below:
- The NGEC can participate in the Interbull truncated-MACE service. By providing reduced-data national EBV to Interbull for truncated MACE, the results returned by Interbull will be the ideal MACE input for reduced-data national genomic evaluation.
- The Interbull Centre can make historical files available upon request, which shall include the official MACE results published 4 years earlier. These MACE proofs will be less ideal than truncated MACE proofs, because current evaluation systems were not re-run with older data by any country for the MACE proofs already computed 4 years earlier.
- The current MACE proofs can be used by excluding all recently proven bulls in MACE who did not have an official MACE proof 4 years earlier. This approach is an exception that should only be used if both preferred options above are impractical. The main concern with this approach is that reduced-data genomic prediction equations will include contributions from phenotypes in most recent 4 years, through sires of the recently proven bulls, and more generally through MACE proofs of all older bull with any relationship to the validation test bulls whose MACE proofs are being excluded.
Specific instructions for data preparation:
- The domestic bulls (type of proof ≠ 21 or 22) that have EDCf ≥ 20 and EDCr = 0 are called test bulls. Test bulls are likely to be included in the genomic reference population with full data, but not with reduced data. Interbull recommends that the reduction in size of the genomic reference population, due to the dropping of test bulls in reduced data, should not exceed 25%
If the size of genomic reference population is reduced by too much, then the accuracy of GEBV calculated from truncated data becomes significantly lower than with full data. In that case, the country can use n<4 years as the time difference between full and reduced data sets.
If the number of test bulls is too small (<50), then the country may choose to also include foreign bulls that have been used locally (type of proof = 21 or 22) with EDCf ≥ 20 local progeny and EDCr = 0 as part of the validation group, to increase the number of test bulls.
- In both exceptions above, the criteria used to define test bulls must be communicated to the Interbull Centre.
- Appropriate time windows (birth years of test bulls) may vary depending on the trait to be validated, the speed of progeny test programs and other factors. The standard adopted for the GEBV test is to include progeny-proven bulls born since (YYYY-8) as test bulls. For instance, if the evaluation year is 2024 and the most recently proven bulls in file300Cf were born in 2020, then the test bulls would include bulls born between 2016 and 2020. Countries may include a wider window of test bulls, or may shift the window by one year, but the reasons must always be communicated to the Interbull Centre.
- Include all available bulls of interest, as described below, in the respective files with their EBVf, EBVr, GEBVf and GEBVr, without editing based on EDCf or EDCr. These final edits, as required for the validation test, are applied within the GEBV test software.
- If the GEBV are a combination of DGV and EBV, then both the DGVr and EBVr used to generate the GEBVr must be estimated from the truncated data.
- Bulls with EBV in the full data sets only, having no progeny information four years ago (EDCr=0), should be included in the reduced-data files (300Cr and 300Gr). Additionally, a minimum of 10 years of bulls with progeny-based EBV in both the full and reduced data sets should be included in the reduced-data files. After recent updates to the software, bulls with progeny in the reduced data are now additionally required as a statistical control group used to improve statistical tests for bias in the evaluations of validation test bulls.
Test description
Testing for bias in the GEBV
Methodology updates in 2024
The official Interbull GEBV test is now based on VanRaden's de-regressed GEBV (described in the 2021 Interbull bulletin paper, https://journal.interbull.org/index.php/ib/article/view/82) as the official prediction target. The VanRaden dGEBV replaces the previously used dEBV target described by Mantysaari et al (2010). Predicting later GEBV or dGEBV from earlier GEBV is conceptually easier to understand and to verify than predictions of dEBV. The new tests are also more suitable for validating single-step models, where genomic preselection effects are properly accounted in GEBVf, GEBVr and dGEBV, whereas dEBV include genomic preselection bias.
With the implementation of a new validation target, the de-regression method has now been internationally standardized because the dGEBV are derived directly from values based on official publication rules, in file300Gf and file300Gr files, in the same way for all countries.
The software will make sure that full and reduced-data evaluations are on the same genetic base of expression by adjusting the mean and variance of reduced-data evaluations to match the base of expression of full-data evaluations. These adjustments to align the evaluation scales are based on bulls already progeny-proven in the reduced data who have expected changes in evaluations very close or equal to zero, due to either no new progeny or relatively few in the recent data. After aligning the evaluation scales, changes in evaluations for the validation test bulls, who have all their progeny in the recent data, are equivalent to contrasts of evaluation changes for validation bulls relative to previous generations of proven bulls who have expected changes of 0.
Average changes in evaluation between reduced and full data will now have an expectation of 0 for any group of bulls, after the scales are aligned. Additional tests have been added, which account for both the combination of intercept and slope estimates from the validation models, to detect probabilities of bias in below-average versus average versus above-average (top) bulls. A new user option to output base-adjusted evaluations from reduced data, for all or selected traits, can also be used to help isolate reasons for detected biases in the evaluations of any traits failing the GEBV test.
Besides the application of the official GEBV test, the software also allows users to choose different validation targets for further internal research. Below is the list of available options:
- file300Df_COUBRD (de-regressed EBV, as used previously in the old test)
- file300Cf_COUBRD (EBV from the full-data evaluation)
- file300Gf_COUBRD (GEBV from the full-data evaluation)
- file300Vf_COUBRD (Any user-defined value, e.g. single step DD, new file)
The user must create whichever input file(s) above are needed for the requested validation target.
A Bootstrapping approach has been implemented to replace the previous t-test for bias in validation slopes, addressing technical concerns that the t-test was not valid, because validation bulls are genetically related, and the validation model residuals are correlated.
The overall validation result, which combines results from either a PASS or FAIL across several sub-tests, will present the following value: PASS, hiSE (i.e. high Standard Error) or FAIL. An overall PASS requires a PASS for the different slope tests plus either a PASS or hiSE for the accuracy test. A result of fail for either the combination of different slope tests or the accuracy test causes an overall FAIL. The new reporting of hiSE indicates too little data to conclusively prove PASS or FAIL in some traits and populations.
Validation regression models
Weighted linear regression models are used to test for bias in both the national genomic and the conventional evaluations, respectively. To pass the official Interbull GEBV test, however, requires only that the GEBVr are unbiased, and not the EBVr. The test for bias in EBVr is provided as comparative and additional information only.
We first define a validation target variable φ that resembles phenotypic progeny averages, and which is based on the progeny contributions in current GEBVf of the validation test bulls. All progeny contributions for the test bulls were from the most recent 4-year period, and contributed to GEBVf and EBVf, but not GEBVr and EBVr. The validation regression models are:
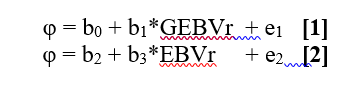
As discussed in the previous section, the validation target φ in the official GEBV test is defined as the dGEBV of VanRaden (2021). The validation test bulls for both models must meet the following criteria: EDCf ≥ 20 and EDCr = 0, born within a pre-defined range of birth years, such as (YYYY-8) to (YYYY-4) inclusive, where YYYY is the current year of evaluation, and having both an EBVr and a GEBVr available. All validation test bulls with an observation in φ are therefore genotyped, and the most recently progeny proven.
The reliability equivalent of information from progeny phenotypes, all of which were included in the full data but not in the reduced data for the test bulls, is used as the regression weight in both models. The progeny information, expressed as an EDC, is first derived from genomic reliabilities based on full (GRELf) and reduced data sets (GRELr), as shown below, and the EDC are then converted back to a reliability equivalent as the bull's regression weight (WT):
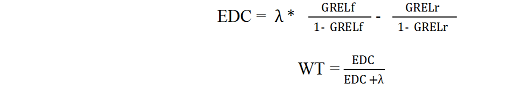
The constant λ is a function of the trait heritability but using any value for λ in the pair of equations above will result in the same WT, so these equations can be simplified by substituting λ=1 in both equations. The WT is thus a function of only GRELf and GRELr.
Effects of selective genotyping
The estimated regression coefficients, b1 and b3 from the two validation models, are compared with expected values to test H0: b1 = E(b1) for GEBVr and H0: b3 = E(b3) for EBVr. The expected values are equal to 1 for both models if all bulls most recently progeny-proven were also genotyped. The expected values will be lower than 1, however, if only a subset of bulls were genotyped, and the genotyped bulls were non-randomly selected with respect to the given trait. The software includes adjustment for the effects of selective genotyping on E(b1) and E(b3).
The first step in deriving the adjustments is to estimate selection differentials for each validated trait. Selection differential is the standardized difference in means between genotyped bulls (g) versus (all) progeny-proven bulls who otherwise qualify as members of the validation test group.
i = (µEBVg - µEBVall)/ σEBVall [3]
Using normal distribution tables from quantitative genetics books (e.g. page 379 from Falconer, D. S. & Mackay, T. F. C. Introduction to Quantitative Genetics, Longman, 4th ed. 1996) the proportion selected by truncation (p) to generate an equivalent selection differential as the observed i, and the corresponding truncation point x that divides the standard normal density into the selected (p) and non-selected (1-p) proportions, can be obtained.
From the equivalent proportion under truncation selection, the expected values of regression coefficients can be approximated using expected effects of truncation selection on the variances and covariance between φ and the independent variable X, where X is either GEBVr or EBVr. Denoting all variables after selection on φ with a superscript s, defining R2b = R2(φ,X) before selection, and following Bulmer (1971) and Henderson (1975):
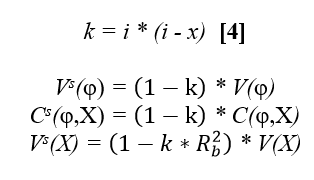
From the expected Cs(φ,X) and Vs(X) after selection, we get the expected b1 after selection:
Es(b1) = ((1 - k) / (1-k*R2b)) * E(b1) [5]
From the observed R2φ,x after selection, with the following expected value, we can derive the required R2b in equation [5] as follows:
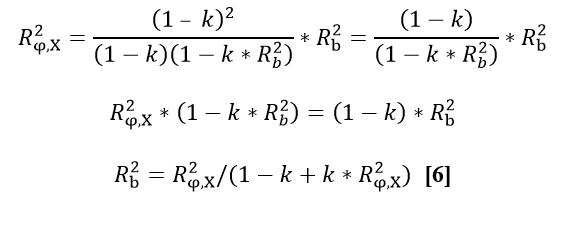
Substituting [6] into [5] and simplifying, we get expected slope as a function of observed R2φ,x
Example: Let µEBVg = 16.00, µEBVall = 11.76, σEBVall = 10.00, R2φ,x = 0.50. Using equation [3], the selection differential (i) for genotyped bulls equals 0.424. For this value of i, the equivalent proportion by truncation selection for genotyping (p) would be 0.75 and the mean deviation of the truncation point from the overall mean (x) would be -0.674 (from reference table). From equation [4] we get k=0.466, and from [6] then [5] we get Rb 2 = 0.652 and then Es (b1) = 0.767, or directly from [7] we also get Es (b1) = 0.767.
Table 1 - Examples of expected regression coefficients (E(b1)) as functions of the selection intensity (i) and the coefficient of determination after selection (R2φ,x ).
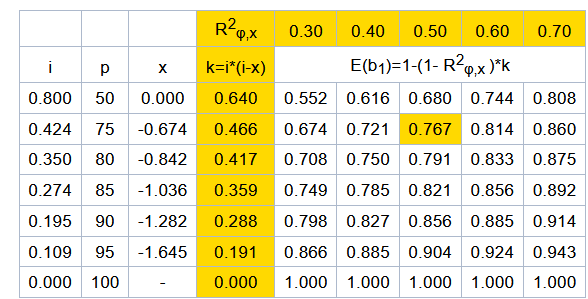
Testing for accuracy improvement with genomics
The improvement in prediction of daughter performance due to the addition of genomic information (i.e. genotyping) is tested by bootstrapping the difference in validation model R2 for models [1] - [2]. A positive difference (P<.05) indicates a significance increase in accuracy with GEBV and therefore results in a Pass. A negative difference (P<.05) results in a Fail, and a non-significant difference (P>.05) indicates that data were insufficient to conclude either way, which therefore results in a designation of hiSE (high standard error). A Pass or hiSE result is required as one part of the overall requirements to PASS the official GEBV test.
Description of National Genomic Evaluations
National Genetic Centres shall provide a description of their national genomic evaluations to Interbull Centre, for now by using the GENO forms, and in the near future, by electronic forms within the PREP database that will be replacing GENO forms.
Updated descriptions shall be provided each time changes to the national genomic evaluations are introduced.
References
Sullivan, P.G. 2023. Updated Interbull software for genomic validation tests. Interbull Bulletin 58, p.7-16.
VanRaden, P.M. 2021. Improved genomic validation including extra regressions. Interbull bulletin 56: 65-69.
Mäntysaari, E., Liu, Z and VanRaden P. 2010. Interbull Validation Test for Genomic Evaluations. Interbull Bulletin 41, p. 17-21.
Bulmer, M.G. 1971. The effect of selection on genetic variability. American Nat. 105:201.
Henderson, C.R. 1975. Best Linear Unbiased estimation and prediction under a selection model. Biometrics 31:423-447.
Falconer, D. S. & Mackay, T. F. C. Introduction to Quantitative Genetics, Longman, 4th ed. 1996
CoP Appendix IX - GMACE Publication Rules
In order to be included in the routine GMACE distribution files, a GMACE record needs to fulfill some conditions related to both inclusion in GMACE calculation process and distribution process.
1-GMACE calculation process
Bull's age <= 7 years
- No distributed conventional proof
- Full pedigree in the Interbull pedigree data base
- Parents with conventional proof in corresponding conventional evaluation or with GMACE evaluation (e.g. young genomic sire of young genomic bull)
- Only bulls with proofs based on parent average +genomic information and transition bulls (those bulls continue getting GMACE until they have enough daughters to qualify for an official MACE proof).
2-GMACE distribution process (Routine run)
- The bull should have at least one GEBV record included in GMACE having a publication status=Y (official in the country of evaluation) and a bull status =10 ( A.I. bull)
- The bull should be authorized to be published by the bull controlling country
- Conflict bulls (claimed by more than one country) will be published if the conflict is not resolved by the countries
- Bulls that are not claimed by any bull controlling country (are not in the 734 file of any country) are considered as publishable if all the other conditions are fulfilled
Bulls that were correctly previously published in GMACE are considered as publishable. The national GE center will have to use the "Exceptions" process (see the diagram below) to This means that a national GE centre will need to write, explain and justify to Interbull centre why a bull that was previously published should not be published anymore.
Clarifications regarding the 734 file sent by countries.
- This file is not mandatory
- All bulls (bulls reported in the previous file and new bulls) need to be included in the 734 file sent by countries.
- If a country choose to send only new bulls:
- There will be no effect on the GMACE publication status of old bulls (reported in the previous file and not in the current one) if they were published before
- However old bulls will not be included in the 734_file_ALL distributed to countries at each run. This file is constructed from all the 734 files sent by countries at each run and start from scratch at each run.
- If a country choose not to send at all any 734 file, new bulls that were not previously published will be published (if all the other conditions above are fulfilled) if no other country include them in its own 734 file with a GMACE publication status =N
CoP Appendix X - AnimInfo
Contents
AnimInfo is a module in the Interbull Data Exchange Area (IDEA) website. The purpose of AnimInfo is to collect reported information from National Genetic Evaluation Centres (NGECs) having access to IDEA and to use AnimInfo as an international exchange area for information on any animals that is present in the IDEA Pedigree module. AnimInfo does not verify or authorize such information. The responsibility of such verification rests with the NGECs and their relevant local/national herdbooks uploading the information. The type of additional information that can be registered in IDEA AnimInfo is decided by Interbull Service Users in collaboration with Interbull Centre.
All data existing within AnimInfo shall remain there and shall continue to be accessible for downloading by any authorised Service User. Data existing within AnimInfo can however be updated by the organisation who submitted the data if it is found to be incorrect.
General Structure
Each additional information recorded via AnimInfo is charaterized by a TYPE, an ATTRIBUTE and a VALUE.
TYPE: The type of information recorded, i.e. Breed Percentage, Genetic Traits etc.
ATTRIBUTE: The attributes holds the actual information of the relevant AnimInfo type, i.e. percentage, genetic codes, population etc.
VALUES: the value of each attribute for each animal. The value must conform to the specification of the attribute, which can be different from attribute to attribute; ranging from a free-form text string, to a set of predefined values, to a defined pattern the value must match.
The system allows different security levels for the information which means that for some AnimInfo information only the authorized organization may view and upload, for other information it is possible for some or all organizations to view and/or upload.
The type of additional information that will be registered in IDEA Animinfo is decided by member organizations in collaboration with Interbull Centre. Interbull Centre will have to register the type of additional information (i.e. coat color, herdbook number etc) in IDEA before member organizations can be able to upload the information via the IDEA Animinfo module. Therefore, member organizations are encouraged to send requests on new additional information types to Interbull Centre.
Quality Control on Uploaded Data in AnimInfo
The AnimInfo file format is an XML file format. For basic information on XML, see https://en.wikipedia.org/wiki/XML or XMLdigest. XML is a flexible system for complex data files and was choosen for AnimInfo in order to ensure easy future development and extension of the module's file format and capabilities, as well as a fitting format for the current data model. To assure that the format of the file is correct and that the information reported are in line with what the module can handle, the data is checked by a checking program developed at the Interbull Centre. The checking program will connect to the IDEA AnimInfo module and check that all the types, attributes and values of information are in line with what specified in AnimInfo. If invalid types, attributes and/or values are detected the program will terminate producing a list of the error detected. In case no problems are found, the program will proceed creating a zip file for uploading. Only information on animals included in the IDEA Pedigree module can be uploaded via AnimInfo.
Correct information depends on the supplier of the information. The Interbull Centre cannot check if the provided information is correct, but does identify information on conflicting information it receives.
Current Type of Information Available via AnimInfo
At the moment, AnimInfo is set up to receive the following additional information for a given animal:
BREED PERCENTAGE: Information about the percent of a breed in a given animal, currently only applied to define the percent of Holstein in Simmental animals.
GENETIC TRAIT: information on genetic traits according to the WHFF, currently only applied to the Holstein breed.
HOL RED TYPE: information to distinguish between B/W and Red Holstein, only applied to the Holstein breed.
Conflicting Information and Conflict Resolution
If conflicting information on a given type of information for the same animal is uploaded or is otherwise publicly available, the conflict needs to be resolved by the NGECs and, if applicable, by the third parties that provided the conflicting information. If they cannot resolve the conflict, they have to contact the bull owner and ask for assistance. Until the resolution of the conflicts both sets of data on the specific animal is disseminated. Following resolution, the correct information should be uploaded as soon as possible. In order to assist with the identification of such conflicts, the NGECs can receive a report from Interbull Centre including all conflicting information for a given type.
CoP Appendix XI - Genetic Traits Exchange
Interbull Centre promotes the collection and facilitates the exchange and conflict resolution for several genetic traits, including recessive traits, via a dedicated module of the Interbull Data Exchange Area (IDEA) called AnimInfo (see Appendix X for more information on how the module works).
The Genetic Traits Exchange service is for the exchange of results from direct genetic test (direct genotyping) of the genetic traits listed for each specific breed. This means that organisations classical haplotype results or indirect test are excluded from the exchange.
Exchange of genetic traits through IDEA/AnimInfo gives several benefits, such as:
- One common platform to share information with other organizations taking part in the service
- Sharing of important genetic information to make better breeding decisions avoiding mating of carriers of recessive diseases or spreading of unwanted alleles in a population.
- As the service is an extension of IDEA pedigree, the consistency of the unique international animal ID is maintained across countries.
- Allowing access to a wider set of information and assuring a smoother and more timely exchange of genetic defects’ information among participating organisations
- Drastically reduce the amount of conflicting information among countries
- Cost reduction by avoiding multiple genetic tests on the same animal for the same traits
Information collected are shared 3 times per year, together with distribution of official international evaluations’ results, to all organisations having signed the specific service agreement (see Appendix I – letter of agreements for genetic traits exchange) and that regularly upload the latest information and actively participate in conflict resolutions.
A minimum requirement for upload is:
- Three times per year, prior to each Interbull Routine Evaluation
- Information on all animals that are included in or contribute to the Interbull Routine Evaluation
The service allows for an easier, safer, smoother and timely exchange of information among participating organisations regarding animals which have been tested carriers of important traits, including recessive traits, and facilitates identification of possible conflicting information and their resolution. Prior to the launch of the Interbull genetic traits exchange service, genetic traits information was exchanged manually and bilaterally between organizations. This was a sub-optimal procedure where often registration of genetic traits was incomplete leading to breeders being often not fully informed thus yielding a problem when carrier animals were mated
If conflicting information on a given type of information for the same animal is detected, the conflict needs to be resolved by the organisations and, if applicable, by the third parties that provided the conflicting information. If they cannot resolve the conflict, they have to contact the bull owner and ask for assistance. Until the resolution of the conflicts, both sets of data on the specific animal is disseminated. Following resolution, the correct information should be uploaded without undue delay. In order to assist with the identification of such conflicts, organisations will receive a report from Interbull Centre including all conflicts found in the system for a given trait.
The service is currently offered for the following breeds: Holstein and Brown Swiss.
Expansion to additional breeds, currently included in an international evaluation, is possible given that a set of standardized coding for the traits of interest is available for the breed at hand. Contact Interbull Centre for more information or for assistance on this step.
The genetic traits exchanged are based on direct genetic test results. The list of traits currently exchanged is reported in Table 1. Full list of codes and explanations are available inside IDEA/AnimInfo.
Table 1: List of traits currently exchanged (NOTE that only direct genetic test results may be uploaded. Haplotype and indirect test results must be excluded):
HOL |
BSW |
BLACK GENE |
ARACHNOMELIA |
BOVINE LEUKOCYTE ADHESION DEFICIENCY (BLAD) |
BH14* |
BLACK/RED GENE |
BH2* |
BRACHYSPINA |
BETA CASEIN |
CHOLESTEROL DEFICIENCY |
FH2* |
CITRULLINEMIA (CIT) |
KAPPA CASEIN |
COMPLEX VERTEBRAL MALFORMATION (CVM) |
OH1* |
DEFICIENCY OF URIDINE MONOPHOSPHATE SYNTHASE (DUMPS) |
ANIMALS BORN HORNLESS (POLLED) |
MULE FOOT |
RENALE DYSPLASIA |
ANIMALS BORN HORNLESS (POLLED) |
SPINAL DYSMYELINATION (SDM) |
RED GENE |
SPINAL MUSCULAR ATROPHY (SMA) |
VARIANT RED GENE |
WDR19 |
FACTOR X1 |
BOVINE PROGRESSIVE DEGENERATIVE MYELOENCEPHALOPATY |
Early Onset Muscle Weakness |
RENAL DYSPLASIA |
* Direct genetic tests, the reason for the naming ‘H' (for Haplotype) come from the fact that these loci were originally detected by Paul VanRaden’s homozygosity depletion approach. But only months later the causative mutations were identified |
|
Participation to the service is subjected to a fee as reported in Table 2. The fee structure will be in place starting from year 2024.
Table 2 – Fee structure for Genetic Trait Exchange:
Exchange of information for: |
Fee (in Euro) |
One Breed |
1 000 |
Any additional breeds |
500 (per breed) |