|
Size: 71148
Comment:
|
Size: 70934
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 170: | Line 170: |
| ^1^Valid record type 300 for EBV and 700 for GEBV <<BR>> <<BR>> ^2^Breed codes accepted: BSW=Brown Swiss type; GUE=Guernsey type; HOL=Holstein-Friesian (Black & White) type; JER=Jersey type; RDC=Red Dairy Cattle type ; SIM=Simmental type.<<BR>> <<BR>> ^3^Valid population codes: ARG AUS BEL CAN CHE ^a^CHR CZE ^b^DEA DEU ^c^DFS ^d^DNR ESP EST FIN FRA ^e^FRM ^f^FRR GBR HUN IRL ISR ITA JPN LTU LVA NLD NZL POL PRT SVK SWE USA URY ZAF <<BR>>where: ^a^Swiss Red Holstein; ^b^ Austria+Germany; ^c^Denmark + Finland + Sweden; ^d^Denmark Red Holstein ^e^ France Montbeliarde; ^f^French Pie Rouge <<BR>> <<BR>> ^4^Accepted traits abbreviations: <<span(style="color: #FF0000;")>>Production<<span>>(mil = milk;fat =fat; pro = protein); <<span(style="color: #FF0000;")>>Conformation <<span>>( sta = stature;cwi = chest width;bde = body depth;ang = angularity;ran = rump angle;rwi = rump width; rls = rear-leg set;rlr = rear-leg rear view;fan = foot angle;hde = heel depth/hoof height; fua = fore udder attachment; ruh = rear udder height; ruw = rear udder width; usu = udder support;ude = udder depth;ftp = front teat placement;ftl = (front) teat length;rtp = rear teat placement;ous = overall udder score; ofl = overall feet&legs score; ocs = overall conformation score; bcs = body condition score; loc = locomotion); <<span(style="color: #FF0000;")>>Udder<<span>> (scs = somatic cell; mas = mastitis);<<span(style="color: #FF0000;")>>Longevity <<span>>(dlo = direct longevity);<<span(style="color: #FF0000;")>>Calving<<span>> (dce = direct calving ease;mce = maternal calving ease;dsb= direct stillbirth;msb = maternal stillbirth);<<span(style="color: #FF0000;")>>Female fertility<<span>> (hco = heifer conception;crc = cow recycling;cc1 = lactating cow's ability to conceive (1);cc2 = lactating cow's ability to conceive (2);int= internval traits);<<span(style="color: #FF0000;")>>Workability <<span>>(msp = milking speed;tem = temperament).<<BR>> <<BR>> ^5^Accepted codes: '''00''' (unknown); '''11''' (based on first crop sampling daughters);'''12''' (based on first and second crop daughters);'''13''' (based on parent average and genomic information only);'''21'''(based on imported semen of proven bull, second crop daughters only); '''22''' (based on mostly, more than 50%, imported daughters or daughters born from imported embryos.) <<BR>> <<BR>> ^6^'''Y''' (if bull proof meets national standards for official publication in the country sending information.); '''P''' (if bull is part of a simultaneous progeny-testing program, but the proof does not yet meet national standards for official publication);'''N''' (otherwise). <<BR>> <<BR>> ^7^Valid codes for status of bulls: '''00'''(unknown);'''10'''(bull randomly sampled through an official AI scheme);'''15''' (young bull, genomically selected);'''20'''(other bull. Records with “20” in this file will be excluded from the international evaluation, unless type of proof is “21”). <<BR>> <<BR>> ^8^Field for number of daughters should be positive. For missing value put 0. <<BR>> <<BR>> ^9^Field for number of herds should be positive. For missing value put 0. <<BR>> <<BR>> ^10^''Production, conformation, udder health, fertility and workability traits:'' Weighting factor used for these traits is “the effective daughter contribution (EDC)”, which is described In the Interbull document Code of practice, Appendix IV, “Weighting factor for international genetic evaluation”, updated April 27, 2004. EDC values should be rounded to the nearest integer value.<<BR>>''Calving:'' The weighting factors used for calving traits it the total number of calvings for the direct effects and number of daughters with calving for maternal effect<<BR>>''Longevity:'' The weighting factor used for longevity traits depends on the national genetic evaluation model. For linear models the weighting factor is the same as described above for conformation, fertility, production, udder health and workability traits. For survival models number of culled daughters is used as the weighting factor.<<BR>> <<BR>> ^11^Reliability values are nationally calculated reliability values expressed in percents with 4 decimials. For missing value put 0.<<BR>> <<BR>> ^12^National predicted genetic merit values published domestically. For threshold models the submitted values are from the underlying scale. For missing values put 9999999999 <<BR>> <<BR>> | . ^1^Valid record type: . 300 for EBV . 700 for GEBV . ^2^Breed codes accepted: . BSW=Brown Swiss type; GUE=Guernsey type; HOL=Holstein-Friesian (Black & White) type; JER=Jersey type; RDC=Red Dairy Cattle type ; SIM=Simmental type. . ^3^Valid population codes: ARG AUS BEL CAN CHE ^a^CHR CZE ^b^DEA DEU ^c^DFS ^d^DNR ESP EST FIN FRA ^e^FRM ^f^FRR GBR HUN IRL ISR ITA JPN LTU LVA NLD NZL POL PRT SVK SWE USA URY ZAF . where: ^a^Swiss Red Holstein; ^b^ Austria+Germany; ^c^Denmark + Finland + Sweden; ^d^Denmark Red Holstein ^e^ France Montbeliarde; ^f^French Pie Rouge . ^4^Accepted traits abbreviations: . Production (mil = milk;fat =fat; pro = protein); . Conformation (sta = stature;cwi = chest width;bde = body depth;ang = angularity;ran = rump angle;rwi = rump width; rls = rear-leg set;rlr = rear-leg rear view;fan = foot angle;hde = heel depth/hoof height; fua = fore udder attachment; ruh = rear udder height; ruw = rear udder width; usu = udder support;ude = udder depth;ftp = front teat placement;ftl = (front) teat length;rtp = rear teat placement;ous = overall udder score; ofl = overall feet&legs score; ocs = overall conformation score; bcs = body condition score; loc = locomotion); . Udder (scs = somatic cell; mas = mastitis);< . Longevity <<span>>(dlo = direct longevity); . Calving (dce = direct calving ease;mce = maternal calving ease;dsb= direct stillbirth;msb = maternal stillbirth); . Female fertility (hco = heifer conception;crc = cow recycling;cc1 = lactating cow's ability to conceive (1);cc2 = lactating cow's ability to conceive (2);int= internval traits); . Workability (msp = milking speed;tem = temperament). . ^5^Accepted codes: .'''00''' (unknown); .'''11''' (based on first crop sampling daughters); .'''12''' (based on first and second crop daughters); .'''13''' (based on parent average and genomic information only); .'''21'''(based on imported semen of proven bull, second crop daughters only); .'''22''' (based on mostly, more than 50%, imported daughters or daughters born from imported embryos.) . ^6^Accepted abbreviations: .'''Y''' (if bull proof meets national standards for official publication in the country sending information.); .'''P''' (if bull is part of a simultaneous progeny-testing program, but the proof does not yet meet national standards for official publication); .'''N''' (otherwise). . ^7^Valid codes for status of bulls: .'''00'''(unknown); .'''10'''(bull randomly sampled through an official AI scheme); .'''15''' (young bull, genomically selected); .'''20'''(other bull. Records with “20” in this file will be excluded from the international evaluation, unless type of proof is “21”). . ^8^Field for number of daughters should be positive. For missing value put 0. . ^9^Field for number of herds should be positive. For missing value put 0. . ^10^ . ''Production, conformation, udder health, fertility and workability traits:'' Weighting factor used for these traits is “the effective daughter contribution (EDC)”, which is described In the Interbull document Code of practice, Appendix IV, “Weighting factor for international genetic evaluation”, updated April 27, 2004. EDC values should be rounded to the nearest integer value.<<BR>> .''Calving:'' The weighting factors used for calving traits it the total number of calvings for the direct effects and number of daughters with calving for maternal effect<<BR>> .''Longevity:'' The weighting factor used for longevity traits depends on the national genetic evaluation model. For linear models the weighting factor is the same as described above for conformation, fertility, production, udder health and workability traits. For survival models number of culled daughters is used as the weighting factor. . ^11^Reliability values are nationally calculated reliability values expressed in percents with 4 decimials. For missing value put 0. .^12^National predicted genetic merit values published domestically. For threshold models the submitted values are from the underlying scale. For missing values put 9999999999 |

USER MANUAL FOR THE IDEA EBV INTERFACE
Preface
The following is a manual to guide the user through the features of the new IDEA. IDEA stands for Interbull Data Exchange Area. IDEA is a restricted area accessible only to member countries through the Interbull website.
Contents
Software
The Software menu gives you access to the Interbull checking programs. By clicking on Software a drop down menu will open and you will be able to choose the type of checking program you are interested in, i.e. Pedigree or Proofs. Under Software you will also find information on the programs and instructions on how to run them.
Proof's Checking Program
A Python program called CheckProofsPara.py will check the 300/700 proof file format and the associated 301/701 parameter file for format correctness against the IDEA EBV User Manual Appendix II. The program prepares a zip file, IB-ORGCODE-IG-yyymmddThhmmss.zip for conventional MACE and IB-ORGCODE-GG-yyymmddThhmmss.zip for GMACE,if no errors are found in the file. The zip file represents your checked data file to upload to the Interbull Centre IDEA database (https://idea.interbull.org/).
The user instructions and file formats (see Appendix I and II) give details on how to run the program and on the checks performed.
Proofs
The Proofs menu gives you access to the main proofs functions which are: Upload, Review, Messages.
Upload and Verify program
By uploading data in IDEA users will no longer need to run the Verify program prior sending data to Interbull Centre. The Verify program will, in fact, be run automatically in IDEA during uploading.
To uploading functionality for parameter and proof files is available under 'Proofs/Upload'. The only file accepted by IDEA is the zip file IB-ORGCODE-XG-yyymmddThhmmss.zip produced by the CheckProofsPara.py, no other files are accepted. Once in 'Proofs/Upload' users will be able to browse the appropriate file and upload it by click on "Submit query".
The uploading of data will not happen on real time but data will be set on a queue and processed on a later time. Right after clicking "Submit query" a message on the screen will display the amount of parameter and proofs records submitted. A confirmation email will be sent to the email address associated to the user that has uploaded data.
The following are the steps the data will go through during uploading:
CheckProofsPara.py will run once again inside IDEA to assure correctness of format and contents
- The Verify program will check your data against the previous one available.
Confirmation email
Either an "EBV upload success" or an "EBV upload failure" email will be sent to you upon completion of the uploading process.
Upload successful
If the uploading is successful the "EBV upload success" email will list some basic information on your data such as:
- Data connected to runid : 1311r
- Number of records read from the parameter file : x
- Number of records read from the proof file : xxxx
- Number of rows in parameterfile associated
- with proofs : x
- Number of flagged rows in proof file : x
- Number of animals found by real AID : xxxxx
- Number of animals found by alias : 0
- Number of animals not present in the pedigree : 0
- Number of records loaded from the file per combination :
- RDC-DEU-cc1: xxx
- RDC-DEU-cc2: xxx
- RDC-DEU-crc: xxx
- RDC-DEU-hco: xxx
- RDC-DEU-int: xxx
The email can contain up to three different WARNINGS. The three first warnings below are associated to attachments:
- Warning: [n] animal(s) were referenced in your file, but not present in the pedigree database. These animals were discarded! See a detailed list in the attached 'missing_animals.csv' file.
- Warning: [n] animal(s) needing updated pedigree records detected! See 'ped_needed.csv' for a complete list of the animals.
- Warning: Use of aliases detected! See 'ped_alias.csv' for a complete list of the animals.
- This warning is generated if the uploaded file contains re-uploadings for a given Breed-Pop-Trait combination which have not been withdrawed first (see Withdraw data):
- WARNING: This dataset re-uploads already existing data, of which some has not been withdrawn first. These combinations have been skipped; the proper way to re-upload combinations is to withdraw them first (or ask IBC staff to reset them). These combinations have been skipped:[......]
An important source of information comes from Number of flagged rows in proof file: this number represents the amount of discrepancies found by the Verify program. If it is 0 (zero) it means that no discrepancies have been found and your data is automatically submitted for the IGE. If it is not 0 (zero) it means that the Verify program has found some discrepancies in your data compared to the previous one available or the system has detected animals with missing pedigree. To double check the data you need to log in IDEA and go to Proof/Review.
Animals not present in IDEA pedigree or lacking pedigree information (i.e. present in the pedigree database but with sire and dam unknown) will be excluded from the international evaluation.
Upload not successful
If the EBV upload fails, you will get this message in the email:
.While uploading your ebv data 'XXX-IG-20131116T185850' an error was encountered. If there are any question, please, contact the Interbull Centre staff for a clarification. NO DATA WERE UPLOADED FROM THIS FILE!
Actions for attachments
Table 1 summarizes the action needed upon reception of a confirmation email with warnings and/or attachments:
Attachments |
Meaning |
Action Required |
Consequences |
missing_animals.csv |
The animals listed in this attachment are not present in IDEA pedigree |
Log in IDEA, |
If pedigree is not provided, animals are excluded from the international evaluation |
ped_needed.csv |
The animals listed in this attachment have sire/dam set to unknown |
If you have pedigree information for these animals: |
If pedigree is not provided, animals are excluded from the international evaluation |
ped_alias.csv |
The animal IDs listed in this attachment are alias IDs |
You are requested to update your own database with the correct animals' IDs. |
Alias IDs are automatically switched to their corresponding official IDs. |
Refer to the section 'Submit/Withdraw data' for more information |
|||
After uploading: What's next?
Uploading represents only the very first step for submitting your data for an IGE. Here is a description of the actions you need to follow to submit your data for an IGE.
Review your data
The Proof/Review page contains all the information you need to review and submit your data for a IGE.
The page contains several important information grouped into three different tables: Filters table, Central table and Actions table
The Filters table allows you to filter and display the information you have uploaded in a way that better suits you. By default the Central table displays all the information associated to your account but you can costumize the displaying of information choosing among:
- A list of the different breeds you have uploaded data for
- A list of the population for which you have uploaded data for
- A list of the traits uploaded
- A list of the different status of the data uploaded
- A list of the data set codes you have uploaded
Clicking "Reset all" will erase all your previous filters and display again all the information associated to your account.
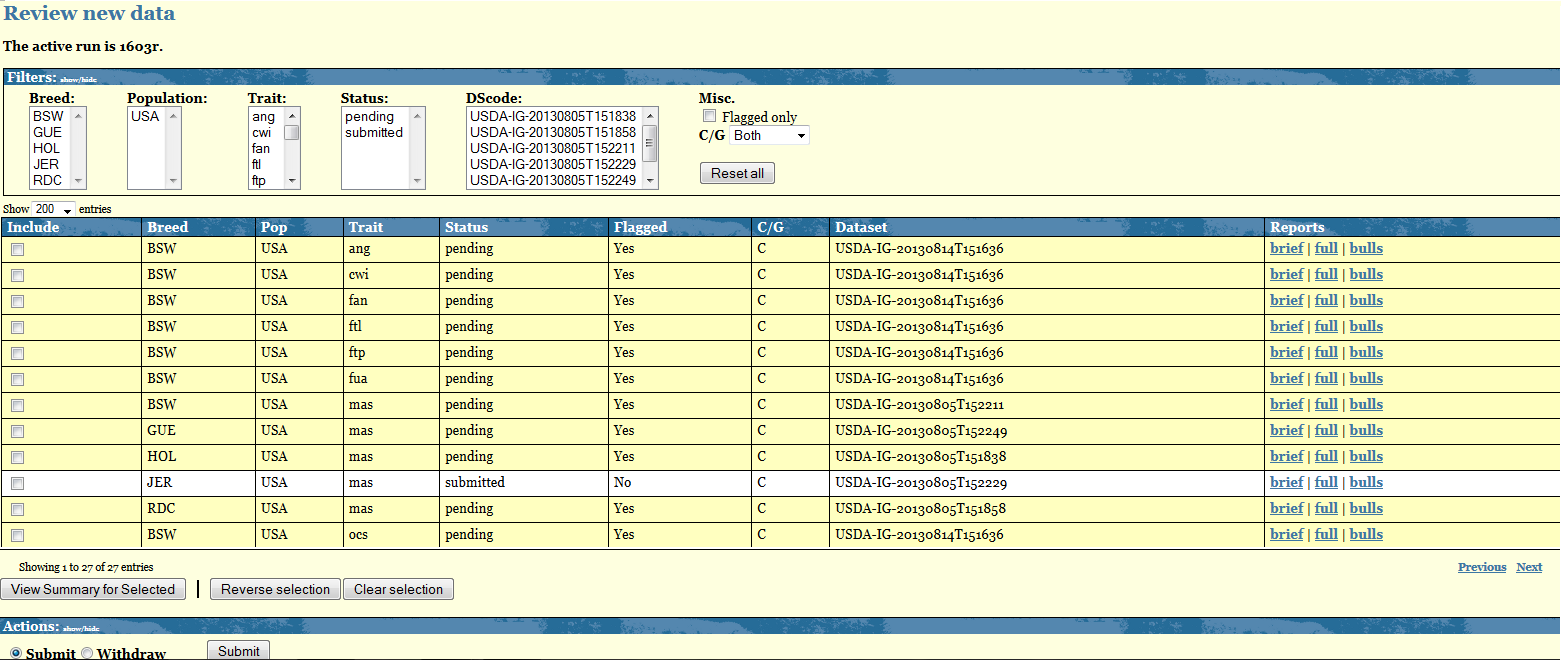
The Central table displays as many rows as the breed-pop-trait combinations you have uploaded. Each row shows the following information:
- Status: refers to the status of your data. There are a total of 5 different status: Pending, Submitted, Withdrawed, Accepted, Rejected
- Flagged: refers to the outcome of the Verify program. Value for this column are either YES or NO. Breed-Pop-trait combination flagged YES are also highlighted in yellow.
- C/G: refers to the nature of your data, Conventional/Genomic. At the moment only Conventional data are accepted.
- Datasets: refers to the dataset used to upload that given Breed-Pop-trait combination
- Reports: for each Breed-Pop-trait combination you get access to a "Brief" and a "Full" output of the Verify program. The "Bulls" report lists all bulls highlighted by the Verify program.
The Breed-Pop-Trait combinations not highlighted and with Flagged=NO are combinations for which the Verify program did not find any discrepancies therefore they get automatically a status=Submitted. No more actions are required from you for these combinations.
The Breed-Pop-Trait combinations highlighted in yellow require your attention. For each of them you need to check the Verify output. For your convenience a "View Summary for Selected" at the end of the Central table will display in a new page the main key checking points of the Verify output. In order for this option to work you need to select some combinations. You can do that either manually by clicking in the box in the "include" column or by clicking on "Reverse selection" and then click on "View Summary for Selected".
The Actions table displays the options you have for your data with status=PENDING. You can decide to submit or withdraw such data by selecting the desired action and clicking on "Submit".
Submit data
If, after checking the Verify output ,you consider your data to be ok and want to include it in the IGE you can do so by selecting the box beside each Breed-Pop-Trait combination you want to include, select the action "Submit" and click on the "Submit" button.
Every time you submit a pending data you are required to explain the reasons for the discrepancies found by the Verify program. If the reason are breed-trait dependent you need to processed these cases one by one and provide the full explanation in the designed space "Change comment". If, on the hand, the same explanation applies to several Breed-Pop-Traits combinations you can select them together in the Review page so that you will be required to write only once the explanation that all these data share.By clicking "Send message" your message will be recorded under IDEA Proofs/Messages and will be visible by you and the Interbull Centre Staff.
In the Review page, the Breed-Pop-Trait combinations you have submitted will be now displayed with status=Submitted.
On the day of the data submission deadline for a given IGE, routine or test run, all your data in the Review page should have status=SUBMITTED. Your aim is therefore to check all pending data and either provide explanations or withdraw and upload new datasets before the data submission deadline.
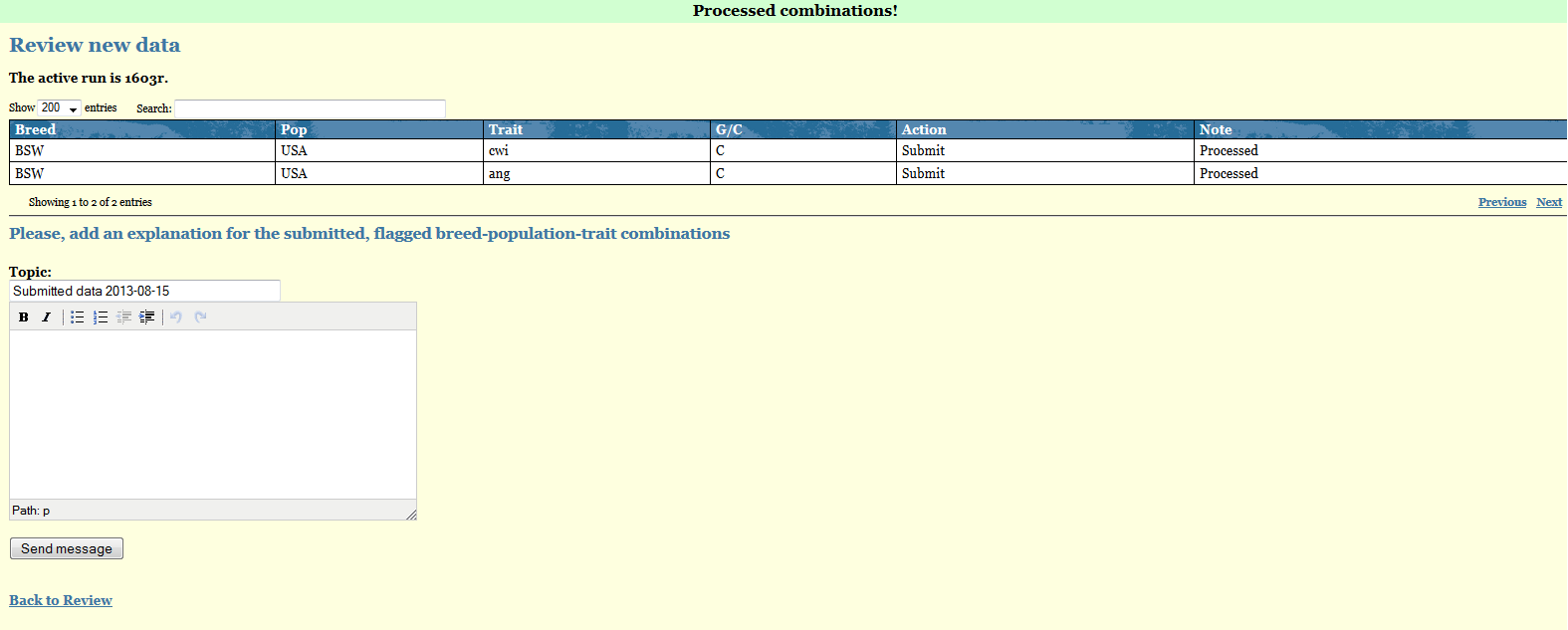
Withdraw data
If you realize the data you have uploaded for some Breed-Pop-trait combinations is wrong or you want to upload pedigree information for the animals reported in the confirmation email attachments you need to withdraw your data before doing anything else. You do so by simply selecting the affected Breed-Pop-Trait combinations in the Review page, select the action Withdraw and then press the button "Submit".In the Review page that given Bredd-Pop-trait combination will now have status=WITHDRAWED.
When you withdraw a given Breed-Pop-Trait all records present in IDEA for that combination are deleted thus you have to re upload the file before the data deadline. It is not acceptable to submit a partial dataset in order to correct the evaluations of some subset of bulls. It is essential that proof records for all bulls be included in the same file for any one brd-pop-trt combination whether the dataset is a first submission for the IGE run in question, or a re-submission with some problem corrected.
If you fail to withdraw all the traits you are going to re-upload in a new file, the confirmation email will contain the following warning:
- WARNING: This dataset re-uploads already existing data, of which some has not been withdrawn first. These combinations have been skipped; the proper way to re-upload combinations is to withdraw them first (or ask IBC staff to reset them). These combinations have been skipped:[......]
Example:
You notice some problem with the temperament data for RDC. You withdraw only the RDC-Tem from your Review table and in the new file to upload you also include data for RDC msp. As you did not withdraw the RDC-msp combination before uploading the new file, all the combinations referring to RDC-msp are skipped as data already exist in IDEA for that given combination.
Accept/Reject data by ITBC. Once you have submitted your data for a given IGE it will be up to the ITBC staff to finally accept or reject it. In general all data automatically submitted and with FLAGGED=NO will also get accepted as there are no issues pending on these data. For all the data with FLAGGED=YES, ITBC staff will go through the explanations you have reported and if found sufficients will accept that given Breed-Pop-Trait combination as accepted. If more clarifications are needed ITBC staff will email you via IDEA, anytime a new message will be posted in your IDEA account by the staff an email will be sent to your email address informing you about the presence of unreplied messages in IDEA.
In case the discrepancies found are considered too big then ITBC staff can reject your data and use the data from the previous run. You will be able to follow what happens to your data by looking at the Review page as the status will change according to what decision has been made.
Messages
Proofs/Messages represents the place where all your communication with the ITBC for a given IGE staff are displayed. All the explanations you provide for a given Breed-Pop-Trait combination are automatically listed in here. Unreplied messages will be marked in red. You can use this page to send further messages to ITBC staff. When ITBC staff replies, their messages will also be listed here and you will be notified by email about the presence of unreplied messages in IDEA.
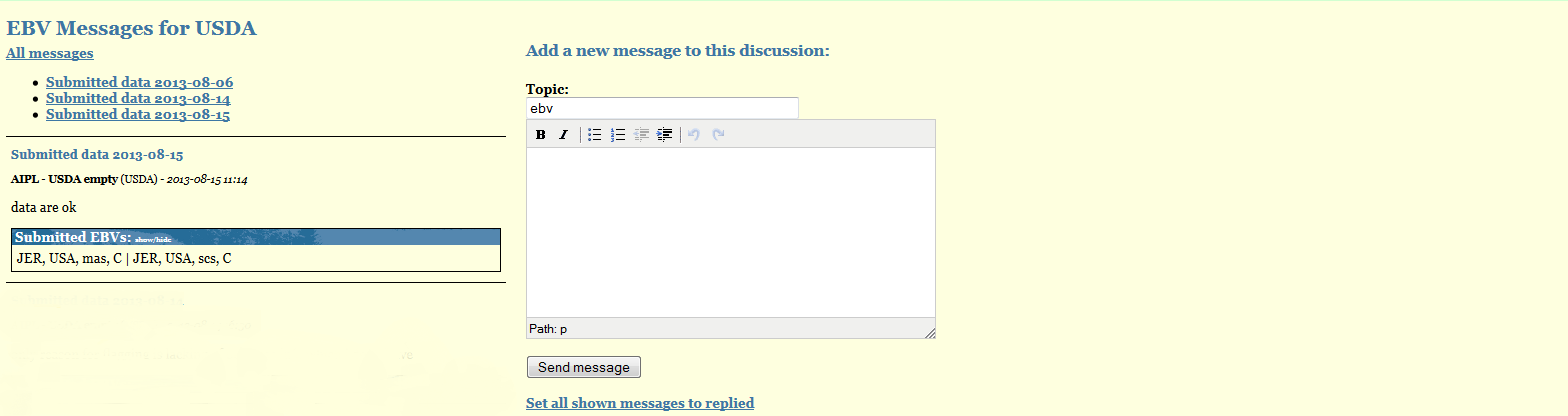
APPENDIX I - Format File300 - Proof file
Col |
Name |
Start |
Format |
Description |
Example |
1 |
rec type |
1 |
a3 |
Record type 1 |
300 |
2 |
brd_eval |
5 |
a3 |
Breed of evaluation 2 |
HOL |
3 |
pop |
9 |
a3 |
Population code 3 |
USA |
4 |
trt |
13 |
a3 |
Trait of evaluation 4 |
mil |
5 |
brd_anim |
17 |
a3 |
Breed of animal |
HOL |
6 |
cou_orig |
20 |
a3 |
Country of first registration |
USA |
7 |
sex |
23 |
a1 |
Sex of animal |
M |
8 |
id_no |
24 |
a12 |
Animal identification number |
840M003000336289 |
9 |
typ_prf |
37 |
i2 |
Type of proof 5 |
11 |
10 |
off_pub |
40 |
a1 |
Official publicationof proof 6 |
Y |
11 |
status |
42 |
i2 |
Animal status 7 |
10 |
12 |
ndau |
44 |
i8 |
Number of daughters 8 |
115 |
13 |
nhrd |
52 |
i8 |
Number of herds 9 |
75 |
14 |
edc |
60 |
i8 |
Number of effective daughter contributions 10 |
133 |
15 |
rel |
69 |
f7.4 |
Repeatability/Reliability 11 |
82 |
16 |
ebv |
76 |
f10. |
National predicted genetic merit 12 |
2.780 |
1Valid record type:
- 300 for EBV
- 700 for GEBV
2Breed codes accepted:
BSW=Brown Swiss type; GUE=Guernsey type; HOL=Holstein-Friesian (Black & White) type; JER=Jersey type; RDC=Red Dairy Cattle type ; SIM=Simmental type.
3Valid population codes: ARG AUS BEL CAN CHE aCHR CZE bDEA DEU cDFS dDNR ESP EST FIN FRA eFRM fFRR GBR HUN IRL ISR ITA JPN LTU LVA NLD NZL POL PRT SVK SWE USA URY ZAF
where: aSwiss Red Holstein; b Austria+Germany; cDenmark + Finland + Sweden; dDenmark Red Holstein e France Montbeliarde; fFrench Pie Rouge
4Accepted traits abbreviations:
- Production (mil = milk;fat =fat; pro = protein);
Conformation (sta = stature;cwi = chest width;bde = body depth;ang = angularity;ran = rump angle;rwi = rump width; rls = rear-leg set;rlr = rear-leg rear view;fan = foot angle;hde = heel depth/hoof height; fua = fore udder attachment; ruh = rear udder height; ruw = rear udder width; usu = udder support;ude = udder depth;ftp = front teat placement;ftl = (front) teat length;rtp = rear teat placement;ous = overall udder score; ofl = overall feet&legs score; ocs = overall conformation score; bcs = body condition score; loc = locomotion);
Udder (scs = somatic cell; mas = mastitis);<
Longevity (dlo = direct longevity);
- Calving (dce = direct calving ease;mce = maternal calving ease;dsb= direct stillbirth;msb = maternal stillbirth);
- Female fertility (hco = heifer conception;crc = cow recycling;cc1 = lactating cow's ability to conceive (1);cc2 = lactating cow's ability to conceive (2);int= internval traits);
- Workability (msp = milking speed;tem = temperament).
5Accepted codes:
00 (unknown);
11 (based on first crop sampling daughters);
12 (based on first and second crop daughters);
13 (based on parent average and genomic information only);
21(based on imported semen of proven bull, second crop daughters only);
22 (based on mostly, more than 50%, imported daughters or daughters born from imported embryos.)
6Accepted abbreviations:
Y (if bull proof meets national standards for official publication in the country sending information.);
P (if bull is part of a simultaneous progeny-testing program, but the proof does not yet meet national standards for official publication);
N (otherwise).
7Valid codes for status of bulls:
00(unknown);
10(bull randomly sampled through an official AI scheme);
15 (young bull, genomically selected);
20(other bull. Records with “20” in this file will be excluded from the international evaluation, unless type of proof is “21”).
8Field for number of daughters should be positive. For missing value put 0.
9Field for number of herds should be positive. For missing value put 0.
10 . Production, conformation, udder health, fertility and workability traits: Weighting factor used for these traits is “the effective daughter contribution (EDC)”, which is described In the Interbull document Code of practice, Appendix IV, “Weighting factor for international genetic evaluation”, updated April 27, 2004. EDC values should be rounded to the nearest integer value.
Calving: The weighting factors used for calving traits it the total number of calvings for the direct effects and number of daughters with calving for maternal effect
Longevity: The weighting factor used for longevity traits depends on the national genetic evaluation model. For linear models the weighting factor is the same as described above for conformation, fertility, production, udder health and workability traits. For survival models number of culled daughters is used as the weighting factor.
11Reliability values are nationally calculated reliability values expressed in percents with 4 decimials. For missing value put 0.
12National predicted genetic merit values published domestically. For threshold models the submitted values are from the underlying scale. For missing values put 9999999999
APPENDIX II - Format File301 - Parameter file
Col |
Name |
Start |
Format |
Description |
Example |
1 |
rec_type |
1 |
a3 |
Record type 1 |
301 |
2 |
brd_eval |
5 |
a3 |
Breed of evaluation 2 |
HOL |
3 |
pop |
9 |
a3 |
Population code 3 |
USA |
4 |
trt |
13 |
a3 |
Trait of evaluation 4 |
scs |
5 |
evdate |
17 |
i8 |
National evaluation date 5 |
20121201 |
6 |
herit |
26 |
f8.6 |
Heritability 6 |
0.12 |
7 |
refbase |
35 |
a7 |
Reference base definition 7 |
H10CB05 |
8 |
pgmdef |
44 |
a2 |
Genetic merit definition 8 |
T- |
9 |
pub_rule |
47 |
a1 |
Official publication rules 9 |
Y |
1Valid record type 301 for EBV and 701 for GEBV
2Breed codes accepted: BSW=Brown Swiss type; GUE=Guernsey type; HOL=Holstein-Friesian (Black & White) type; JER=Jersey type; RDC=Red Dairy Cattle type ; SIM=Simmental type.
3Valid population codes: ARG AUS BEL CAN CHE aCHR CZE bDEA DEU cDFS dDNR ESP EST FIN FRA eFRM fFRR GBR HUN IRL ISR ITA JPN LTU LVA NLD NZL POL PRT SVK SWE USA URY ZAF
where: aSwiss Red Holstein; b Austria+Germany; cDenmark + Finland + Sweden; dDenmark Red Holstein e France Montbeliarde; fFrench Pie Rouge
4Accepted traits abbreviations: Production(mil = milk;fat =fat; pro = protein); Conformation ( sta = stature;cwi = chest width;bde = body depth;ang = angularity;ran = rump angle;rwi = rump width; rls = rear-leg set;rlr = rear-leg rear view;fan = foot angle;hde = heel depth/hoof height; fua = fore udder attachment; ruh = rear udder height; ruw = rear udder width; usu = udder support;ude = udder depth;ftp = front teat placement;ftl = (front) teat length;rtp = rear teat placement;ous = overall udder score; ofl = overall feet&legs score; ocs = overall conformation score; bcs = body condition score; loc = locomotion); Udder (scs = somatic cell; mas = mastitis);Longevity (dlo = direct longevity);Calving(dce = direct calving ease;mce = maternal calving ease;dsb= direct stillbirth;msb = maternal stillbirth);Female fertility (hco = heifer conception;crc = cow recycling;cc1 = lactating cow's ability to conceive (1);cc2 = lactating cow's ability to conceive (2);int= internval traits);Workability (msp = milking speed;tem = temperament).
5 National evaluation dates expressed using the format YYYYMMDD
6Heritability for a specific trait in format f8.6. Should be larger than 0 and smaller than 0,999999
7 Reference (genetic) base definition in the country sending information: breed initial (1 char), year established (YY), bull(b) or cow(C) (1 char); birth(B), calving (C) or evaluation (E) (1 char); year of event (use middle year if base on multiple year (YY); for breed initial see breed code in footnote2 (use X If based on multiple breeds):e.g. H00BB95 means a base defined 2000 base on Holstein bulls born in 1995.
8Genetic merit definition consists of a letter and a sign. B = Breeding value, T = Transmitting ability; Sign: ‘+’ = Higher values are desirable, ‘-‘= Lower values are desirable
9 Allowed characters Y=Yes or N=No
APPENDIX III - Frequently Asked Questions
My data has been rejected, what can i do?
National data can sometimes be rejected from ITBC staff if changes in sire standard deviations between previous and current data are larger than 5% (in case of routine run), or if problems in the data are found. If your data get rejected it means that the previous available data will be used for the IGE. In case you are able to fix the problem(s), and the data deadline has not yet passed, you can try to upload a new corrected dataset. You will need to check the conformation email, the verify output and submit the data in the Review page again.
I discovered a problem with a dataset that got automatically submitted, what do I do?
Inform ITBC staff about the problem and the breed-pop-trait combination affected by it. If the data submission deadline has not yet passed, ITBC staff will reset your data so that you will be able to upload a new file again. You will need to check the conformation email, the verify output and submit the data in the Review page again.
